Figure 1.
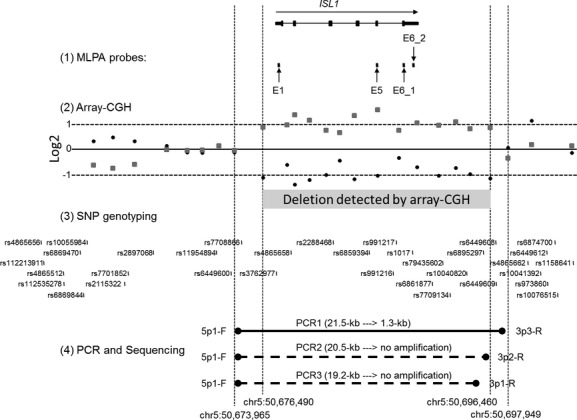
Genomic organization of ISL1 and 20-kb deletion. The genomic organization of the six exons of human ISL1 is shown at the top of the Figure. (1) Four MLPA probes were designed to amplify exons 1, 5, and 6 and are designated by arrows as E1, E5, E6_1, and E6_2, respectively. Each exon showed a loss of copy number by MLPA (data not shown). (2) Two array-CGH experiments with Agilent 1 million features microarrays were performed: hybridization with Cy3-labeled test versus. Cy5-labeled reference DNA (Log2 values plotted by black circles) and Cy5-labeled test versus. Cy3-labeled reference probes (“dye swap” whose Log2 values are plotted by gray squares). The Log2(Test/Reference) values were plotted for each microarray oligonucleotide feature at each chromosomal locus. Negative Log2 values of 14 consecutive probes (black circles) are seen in a zoomed-in array-CGH plot encompassing ISL1 on chromosome 5, indicating a deletion. A nearly mirror image of these Log2 values is observed from the dye-swap experiment (gray squares). The two outside dashed vertical lines indicate the positions of the innermost oligonucleotide features that showed normal Log2 values. The two inner dashed vertical lines show the positions of the two outermost of the 14 oligonucleotide features that showed negative Log2 values. (3) Thirty-four genotyped SNPs are shown relative to the genomic organization of ISL1. (4) We designed several PCRs to identify the chromosomal breakpoints. PCR1: a 21.5-kb product would be expected from the normal chromosome 5 using a primer set, 5p1-F and 3p3-R, indicated by black circles, and whose sequences are listed in Table 2. A 1.3-kb amplicon was generated (see also Figure 2 lane 2) because these primers were brought into proximity of each other by the deletion of the intervening 20-kb chromosomal segment. PCR2 and 3: No PCR product was generated using the same forward primer (5p1-F), in combination with two different reverse primers, 3p2-R and 3p1-R, located inside the right pair of vertical dashed lines. This indicates that these two reverse primers are located within the deleted region.
