Figure 1.
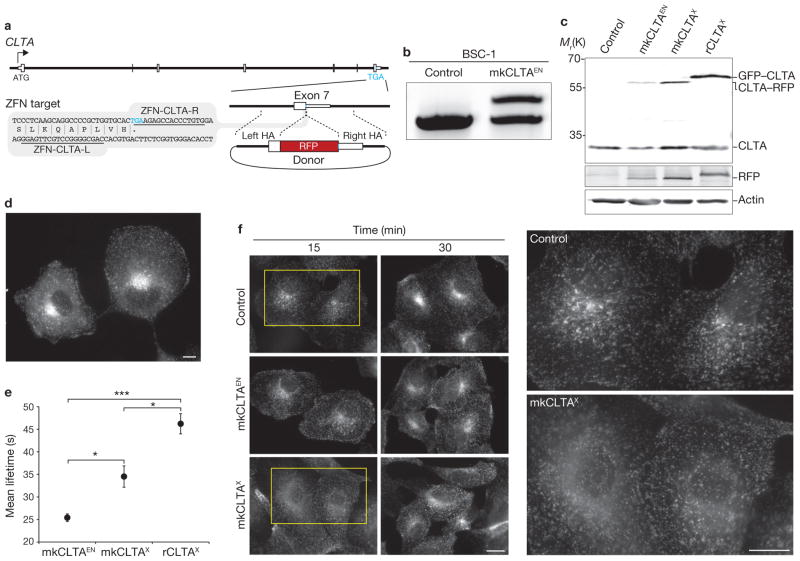
Editing of CLTA using ZFNs in BSC-1 cells. (a) Schematic representation of the strategy for integration of RFP at the CLTA locus. White boxes, exons of CLTA; HA, donor plasmid region of homology to CLTA sequence; Blue letters, stop codon. The grey box indicates the region of CLTA exon 7 surrounding the translation stop codon; the underlined sequences indicate the recognition stretches of the individual zinc finger nucleases (ZFN-CLTA-L and ZFN-CLTA-R, respectively). (b) Out-out PCR showing targeted integration of RFP. Control, parental cell line; mkCLTAEN, single-allele CLTA–RFP tagged genome-edited line. (c) Western blot analysis of cell lysates immunoblotted for CLTA, RFP and actin. Note RFP antibody cross-reactivity with GFP. mkCLTAX, stable CLTA–RFP overexpression line; rCLTAX, stable GFP–CLTA (rat brain-derived) overexpression line. (d) Epifluorescence image of mkCLTAEN cells expressing endogenous CLTA–RFP. Scale bar, 10 μm. (e) CCP dynamics were assessed in the indicated BSC-1 cell lines by quantifying the lifetime of fluorescently tagged CLTA proteins at the plasma membrane. Data are means ± s.e.m. Tracks, 30,734–50,250; n = 5, 11 and 15 cells for the mkCLTAX, mkCLTAEN and rCLTAX lines, respectively. Triple asterisk indicates P < 0.0001 and single asterisk indicates P < 0.05. (f) Time course of Alexa Fluor 488-conjugated human transferrin uptake in the indicated BSC-1 cell lines. Areas enclosed by yellow boxes are enlarged (right) for better visualization. Scale bar, 10 μm. Uncropped images of gels and blots are shown in Supplementary Fig. S2a.