Fig. 2.
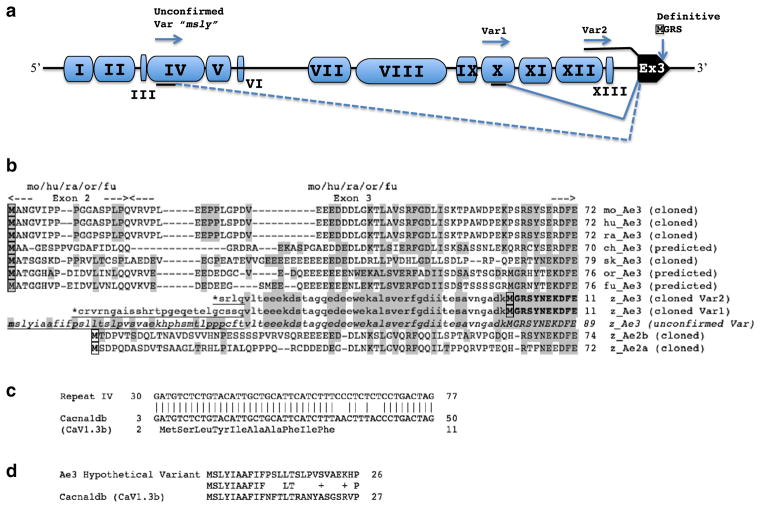
Repeat element insertions at the 5′-end of the zebrafish ae3 gene. a. Schematic of the ~7 kb region encompassing the 5′-end of the zebrafish ae3 gene and upstream sequence. Blue pods labeled with black roman numerals represent repeat elements of distinct sequence; the black pod is ae3 Exon 3. The number of imperfect copies (nt identities up to 90 %) detected in the zebrafish Zv9 genome for each (otherwise unannotated) repeat type (nt numbering from FP243360) was 107 copies of repeat I (2660–2947); 159 copies of repeat II (2946–3545); 100 copies of repeat III (3635–3727); 181 copies of repeat IV (3739–4392); 111 copies of repeat V (4393–4545); 103 copies of repeat VI (4585–4655); 189 copies of repeat VII (5706–6266); 153 copies of repeat VIII (6366–7467); 106 copies of repeat IX (7652–7859); 36 copies of repeat X (7875–8228); 122 copies of repeat XI (8230–8535); 496 copies of repeat XII (8550–9185); 116 copies of repeat XIII (9188–9284). The MSLY Met codon of hypothetical transcript Var3 is located at nt 3760–2 within repeat IV (black underline), and is predicted to be spliced to Ex3 (dashed splice line). The 5′-most extent of validated transcript Var1 within repeat X encodes an in-frame termination codon (*) in an exonic sequence (black underline) spliced to Ex3 (solid splice line). The 5′-most extent of validated transcript Var2 is within repeat XII, and the transcript is continuous (black overline) through the rest of putative ae3 intron 2 (including repeat XIII) into Ex3. Transcript variants 1 and 2 both encode the definitive (confirmed) initiator Met in Ex3 (boxed) starting the polypeptide presented in Figs. 1 and 2. Inter-repeat distances are not to scale. b. Aligned N-terminal Ae3 amino acid sequences (definitive or predicted, as indicated) from zebrafish, mouse, human, rat, chicken, skate, fugu, and tilapia, and from zebrafish Ae2a and Ae2b. The exon 2–3 junction shown at top applies to mouse, rat, and human sequences, and to the predicted sequences of tilapia and fugu. Highlighted in gray are zebrafish Ae3 residues shared in all or some of the other N-terminal sequences. Lowercase residues indicate potential zebrafish Ae3 Ex3 coding sequences confirmed by RT-PCR. The underlined lowercase residues (N-terminally marked with *), RT-PCR proven exonic sequences upstream of zebrafish Ex3, encode in-frame terminator codons and therefore represent 5′-UTR. The third instance of underlined lowercase residues (in italics) is encoded upstream of Exon 3 by a hypothetical exonic sequence remotely homologous to authentic or predicted N-terminal protein coding regions in ae3 genes of tilapia (Oreochromis niloticus; or, XP_003443372), skate (sk, CAD61187), fugu (fu, XP_003962174) chicken (ch, XP_003641688), as well as of mouse (mo, NP_033234), rat (ra, NP_058745), and human (hu, NP_005061). c. Aligned nucleotide sequences of Repeat IV from the zebrafish ae3 5′-upstream region and the N-terminal open reading frame of zebrafish cacna1db (AY528225) encoding the pore subunit of an L-type Ca2+ channel. d. Aligned N-terminal amino acid sequences encoded by hypothetical zebrafish ae3 transcript variant “msly” and by the zebrafish cacna1db gene encoding the Cav1.3b pore-forming subunit of the L-type voltage-gated Ca2+ channel (AAS20587). The shared N-terminal sequences are both encoded by exonized variants of repeat IV