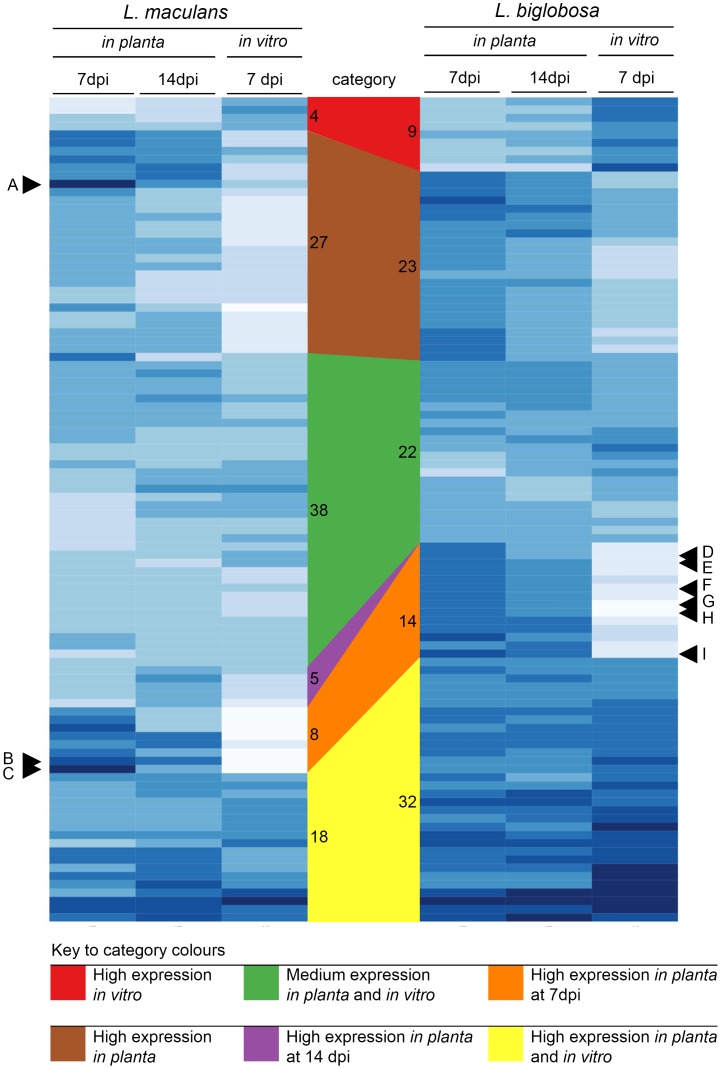
Figure 4. Expression profiles of secreted CAZys of L. maculans ‘brassicae’ (Lmb), L. biglobosa ‘canadensis’ (Lbc) in planta and in vitro.
The top 100 genes expressed across the three treatments (7, 14 dpi and in vitro) and predicted to be secreted and to contain a CAZy domain (CBM, GH, PL, or AA) were selected. Quantile-normalisation was applied and log10-transformed FPKM values were graphed. The intensity of blue shading is proportional to the expression level. The gene order is based on a dendrogram created from a Euclidean similarity matrix with average group distance. Letters with a triangle point to genes that were amongst the top 20 most highly expressed genes in vitro or in planta (Tables 2, 3, 4, or 5). A/Lema_P102640.1 (Lm2LysM); B/Lema_P013700.1 (Glycoside hydrolase family 7); C/Lema_P070100.1 (Lm5LysM); D/Lb_j154_P005286 (pectate lyase); E/Lb_j154_P009204 (cellulase); F/Lb_j154_P001246 (Glycosyl hydrolase family 43); G/Lb_j154_P001652 (cellulase); H/Lb_j154_P009247 (Glycosyl hydrolases family 12); I/Lb_j154_P004093 Glycosyl hydrolases family 39). Categories of gene expression (High expression in vitro, High expression in planta, Medium expression in planta and in vitro, High expression in planta at 14 dpi, High expression in planta at 7dpi, High expression in planta and in vitro) were manually assigned and indicated by coloured polygons linking the genes in each category across Lmb and Lbc; the number of genes in each category is indicated on the vertical sides of the polygon.