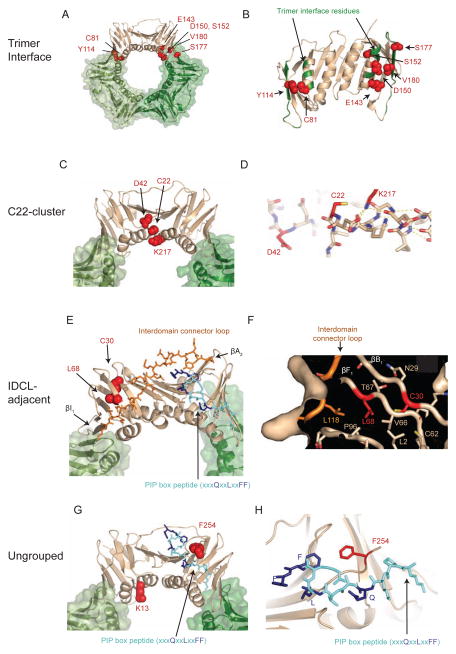
Figure 1. pol30 mutations that disrupt Exo1-independent MMR cause amino acid changes that form clusters on the PCNA crystal structure.
(A–B) The positions of amino acids altered by trimer interface mutations. (A) Mutated amino acids are depicted as red spheres on the tan subunit in the PCNA trimer. (B) Rotated view of the PCNA monomer displays mutated residues as red spheres. Other non-mutated trimer interface residues are colored green on the backbone ribbon. (C–D) The positions of amino acids altered by the C22-cluster mutations. (C) Displayed as in (A). (D) The K217 side chain (red) caps the C-terminus of the αA1 helix by hydrogen bond interactions with backbone carbonyls (dashed yellow lines) and may help position C22 (red). (E–F) The positions of the amino acids altered in the IDLC-adjacent mutations. (E) Displayed as in (A) with the IDCL as orange sticks and the PIP-box peptide as blue sticks, with conserved residues as dark blue. Positions of the two β-sheets (βI1 and βA2) at either end of the IDCL are labelled. (F) Cutaway surface depiction shows part of the hydrophobic core containing L68 and C30 that pack with residues of the IDCL and another β-sheet that interacts with the adjacent PCNA protomer. (G–H) The positions of the amino acids altered in the unclustered mutations. (G) Displayed as in (A) with the PIP-box peptide as blue sticks, with conserved residues as dark blue. (H) Position of the F254 side chain (red sticks) relative to the PIP-box peptide (blue sticks). PCNA and PCNA/PIP-box peptide structures rendered using PDB entries 1plq and 2od8 (Krishna et al., 1994; Vijayakumar et al., 2007). Also see Figure S1.