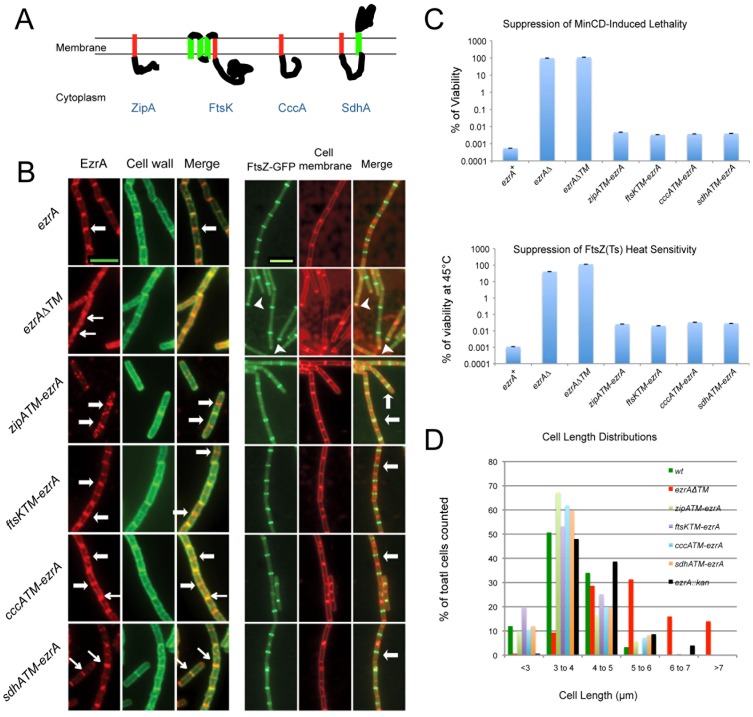
Figure 3. Swapping EzrA's transmembrane helix with similar domains from other proteins does not impact EzrA localization or function.
(A) Cartoon of proteins used to generate EzrA TM chimeras. The TM employed in the chimera is highlighted in red. (B) (Left) Immunofluorescence images of cells expressing wild type EzrA, an EzrA deletion mutant missing the entire transmembrane domain, as well as four EzrA chimeras in which the transmembrane domain has been substituted with the appropriately oriented TM from a heterologous protein. EzrA localization is wild type in all four chimeras. (Right) Localization of FtsZ-GFP in ezrA-TM chimera cells. FtsZ localization is wild type in all four chimeras. Thick arrows indicate medial EzrA and FtsZ localization. Thin arrows indicate EzrA localization at septa. Arrowheads indicate polar FtsZ rings. Exposure times are equivalent for each fluorophore. Bars = 5 µm. (C) All four chimeras behave like wild type ezrA alleles with regard to their sensitivity to MinCD overexpression and heat sensitivity in an ftsZts background. Bars equal standard deviation from three repeated experiments. (D) A plot of cell length distributions between wild type and mutant strains indicates that cell length is not impacted by the TM swaps.