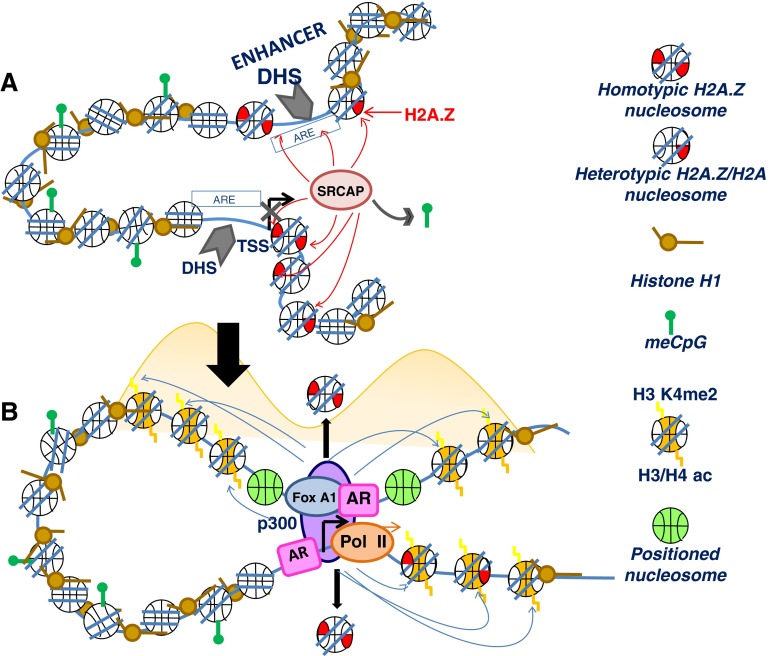
Fig. 2.
The enhancer and promoter regions come in close proximity during the process of gene activation in AR regulated genes. a The process involves SRCAP, a well known co-activator of AR responsible for the deposition of H2A.Z–H2B dimers into chromatin. SRCAP is an integral part of a large ATP-dependent chromatin remodeling complex which contains, among other subunits, the histone acetyltransferase p300/CBP and the forkhead protein FoxA1. The resulting complex brings RNA pol II to the transcriptional start site. Notice that in this proposed model, incorporation of H2A.Z results in histone H1 exclusion [93], demethylated DNA, and nucleosome depleted regions [106] that are responsible for the presence of DHS domains [100, 101]. b The activation model proposed is based on several observations from different groups and includes: presence of two positioned nucleosomes (shown in green) at AR regulated enhancers [102, 103] that are brought in close proximity to the promoter by chromatin looping [111], a process that likely involves FoxA1 [112]. These nucleosomes are flanked by p300/CBP acetylated chromatin domains spanning approximately 2,000 base pairs [102, 103]. The shadowed yellow waves denote the distribution of histone acetylation upon AR induction of gene expression