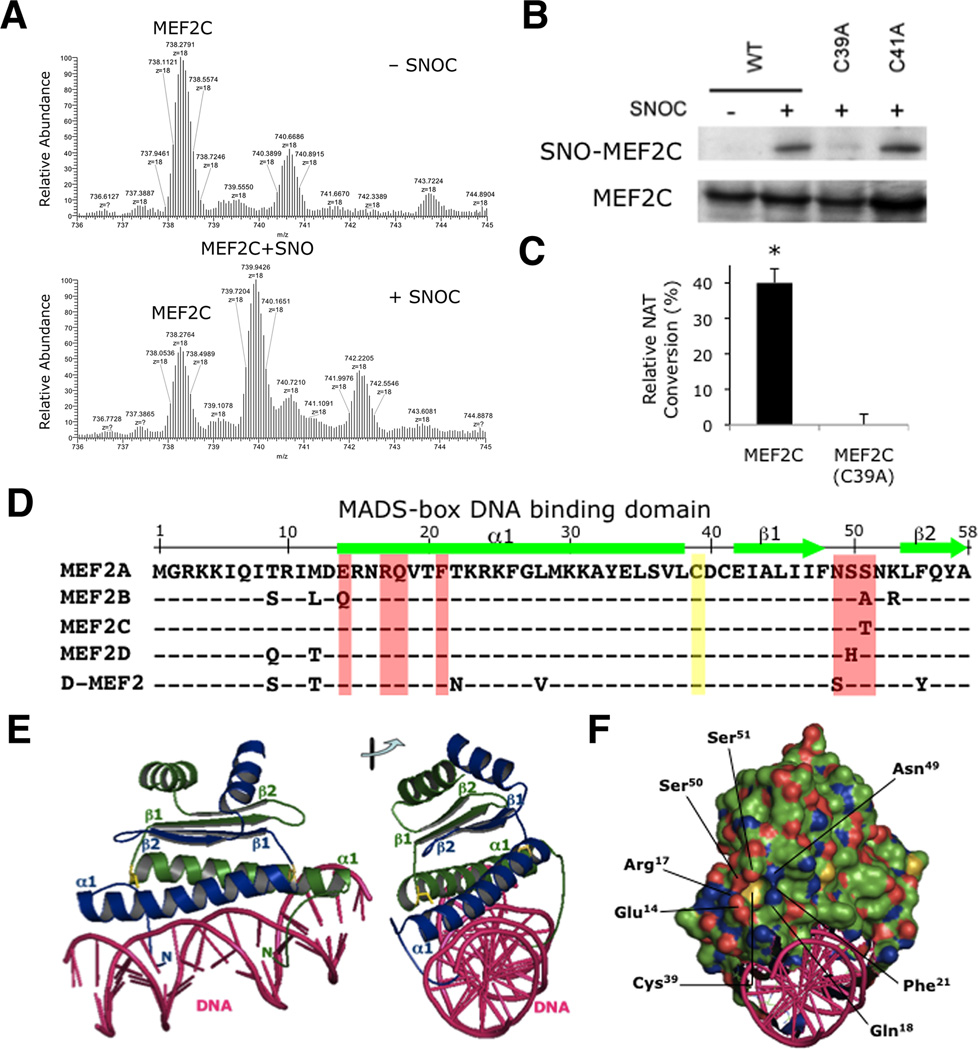
Figure 2. S-Nitrosylation of MEF2 at Cys39 in the MADS-Box DNA Binding Domain.
(A) Recombinant MEF2C protein was infused into a mass spectrometer (LTQ-Orbitrap-XL with ETD) to obtain molecular ion spectra in the presence or absence of NO donor, 50 µM SNOC (n = 2 independent experiments).
(B) HEK293T cells transfected with WT myc-tagged MEF2C (WT), MEF2C(C39A) or MEF2C(C41A) were incubated with SNOC. SNO-MEF2C was determined by biotin switch. SNO proteins were precipitated and SNO-MEF2C detected with anti-myc antibody (n = 3 independent experiments; quantification shown in Figure S2A).
(C) HEK293 cells stably expressing nNOS were transfected with V5-tagged MEF2C (WT) or V5-tagged MEF2C(C39A), and nNOS activated with Ca2+ ionophore A23187. V5-tagged MEF2C proteins were immunoprecipitated with anti-V5 antibody and subjected to DAN assay. Values are mean + SEM (n = 3 independent experiments, *p < 0.05 by t test).
(D) Sequence alignment of MADS-box DNA binding domains of MEF2 (human isoforms MEF2A-D; Drosophila D-MEF2). α-Helix (bars) and β-strands (arrows). Cys39 S-nitrosylation site (yellow). Residues involved in pocket formation (red).
(E) Cys39 (yellow) is symmetrically located at the hinge between α1-helix and β1-strands in the MEF2 (monomer A in blue, monomer B in green)-DNA (pink) complex (PDB ID: 1EGW). Right: viewed at 90°.
(F) Electrostatic view of MEF2 (negative charge in red, positive charge in blue). Cys39 is present at the bottom of the pocket created by Glu14, Arg17, Ser50 Ser51, Asn49, Phe21, and Gln18.
(G) Cys39 of MEF2 monomer A (blue) is surrounded by residues (Glu14 Arg17, Ser50, Ser51, Asn49, Phe21, and Gln18; red) of monomer B (green). Right: complex viewed at 90°.
See also Figure S2.