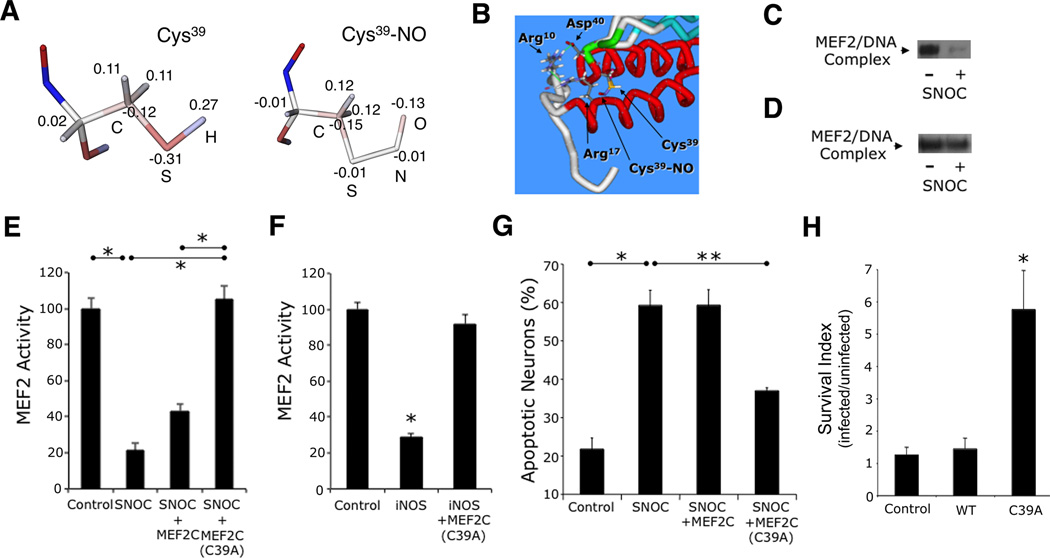
Figure 3. S-Nitrosylation of MEF2 Impairs its DNA Binding, Transcriptional, and Pro-Survival Activities.
(A) Computational structure analysis of S-nitrosylated Cys39. Atomic charge distribution for normal Cys and Cys-NO residues obtained from quantum mechanical calculations (see text). Electrostatic potentials: positive charge (blue) and negative charge (red). Values show excess atomic charge.
(B) Optimization of molecular mechanical geometry to show possible orientation of the Cys-NO sidechain in MEF2. Key residues of dimer interface are highlighted.
(C) EMSA of MEF2/DNA-binding activity. Nuclear extracts from cortical neurons were exposed to 50 µM SNOC (n = 3 independent experiments). MEF2/DNA-binding complex (arrow).
(D) EMSA of mutant MEF2/DNA-binding activity. In vitro translated MEF2C(C39A) was exposed to SNOC (n = 3 independent experiments).
(E) Effect of NO on MEF2 reporter gene activity. Cortical neurons were transfected with empty vector (pcDNA 3.1) or expression vectors for MEF2C or MEF2C(C39A), MEF2 luciferase reporter construct, and renilla luciferase vector to control for transfection efficiency. Luciferase activity was measured 4 h after 50 µM SNOC exposure. Bars represent mean + SEM (n = 12 cultures derived from 3 independent platings, *p < 0.0001 by ANOVA with post-hoc Scheffé's test).
(F) Effect of iNOS transfection on endogenous MEF2 reporter gene activity. Cortical neurons were transfected with empty vector (pcDNA 3.1) or expression vectors for iNOS with or without MEF2C(C39A), and MEF2 luciferase reporter construct plus renilla luciferase; 24 h after transfection, luciferase activity was measured. Bars represent mean + SEM (n = 12 cultures derived from 3 independent platings, *p < 0.0001 by ANOVA with post-hoc Scheffé's test).
(G) Effect of MEF2 S-nitrosylation on neuronal apoptosis. Cortical neurons were transfected with control vector, MEF2C, or MEF2C(C39A) plus enhanced green fluorescent protein (EGFP) vector to identify transfected neurons. Cultures were then exposed to 50 µM SNOC and analyzed 8 h later. Hoechst DNA dye was used to assess condensed, apoptotic nuclei. Bars represent mean + SEM (n = 9 cultures derived from 3 independent platings, *p < 0.01, **p < 0.05 by ANOVA with post-hoc Scheffé's test). (H) Stereotactic injection of S-nitrosylation-resistant MEF2C(C39A) mutant (LV-C39A) but not WT MEF2C into striatum improved neuronal survival in stroke penumbra. Values are mean + SEM of survival index, representing the inverse of the apoptotic ratio for infected vs. uninfected neurons for each construct (n ≥ 4 animals for each condition, *p < 0.05 by ANOVA with post-hoc Scheffé's test).
See also Figure S3.