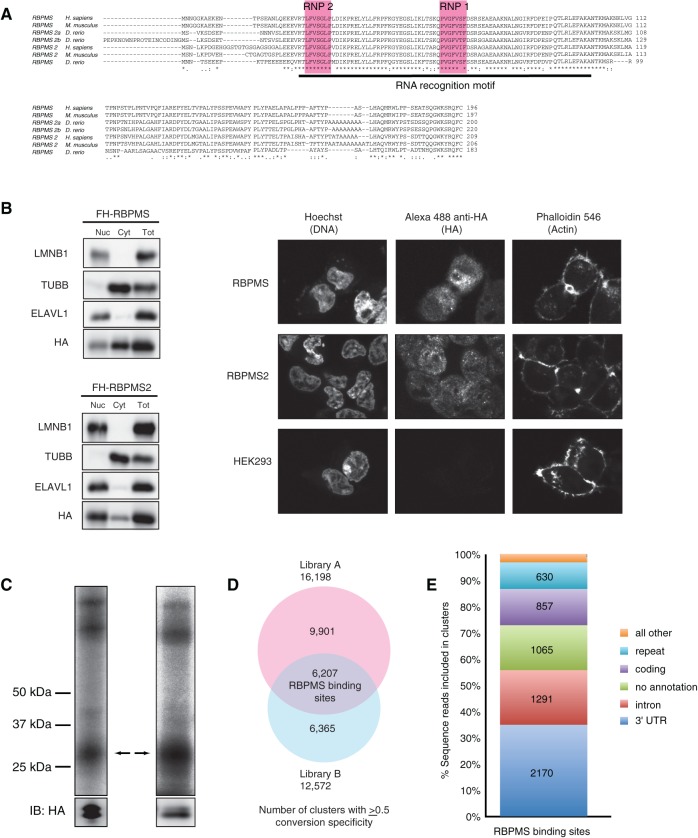
FIGURE 1.
RBPMS overview. (A) RBPMS is conserved in vertebrates. Alignments were generated using ClustalW (NP_956553.1, NP_001002409.1, NP_082306.2, NP_919248.1, NP_001008710.1, NP_001036139, XP_003199078.1). (B) RBPMS and RBPMS2 localize to the nucleus and cytoplasm. Western Blotting analysis of nucleo-cytoplasmic fractions prepared from FLAG-HA RBPMS and RBPMS2 cell lines, as indicated. Nuclear (Nuc), cytoplasmic (Cyt), and total cell lysate (Tot) fractions were resolved on a 4%–12% SDS–polyacrylamide gel and then probed using an anti-HA antibody targeting FLAG-HA RBPMS and RBPMS2, and antibodies targeting ELAVL1 (HuR), TUBB (β-tubulin), and LMNB1 (lamin B) as controls for the purity of the cytoplasmic and nuclear fractions. (Right) Immunofluorescence staining of HA epitope in HEK293 cells stably expressing FLAG-HA RBPMS and FLAG-HA RBPMS2, using Hoechst and Phalloidin 546 as controls. (C) Phosphorimages of SDS-PAGE fractionating PAR-CLIP immunoprecipitate from constitutive and inducible overexpressing FLAG-HA-tagged RBPMS HEK293 cells. The cross-linked RNA–RBPMS complexes are indicated for two biological replicate experiments. Anti-HA Western-blotting control for expression and loading is shown at the bottom. Two protein bands were recognized by anti-HA antibody, one at the expected size based on recombinant full-length RBPMS and the other shorter, suggesting proteolytic cleavage (bands confirmed as RBPMS by mass spectrometry). (D) Overlap of PAR-CLIP clusters with ≥0.5 T-to-C conversion specificity between libraries A and B defines 6207 RBPMS-binding sites. (E) Genomic distribution of RBPMS-binding sites.