FIGURE 5.
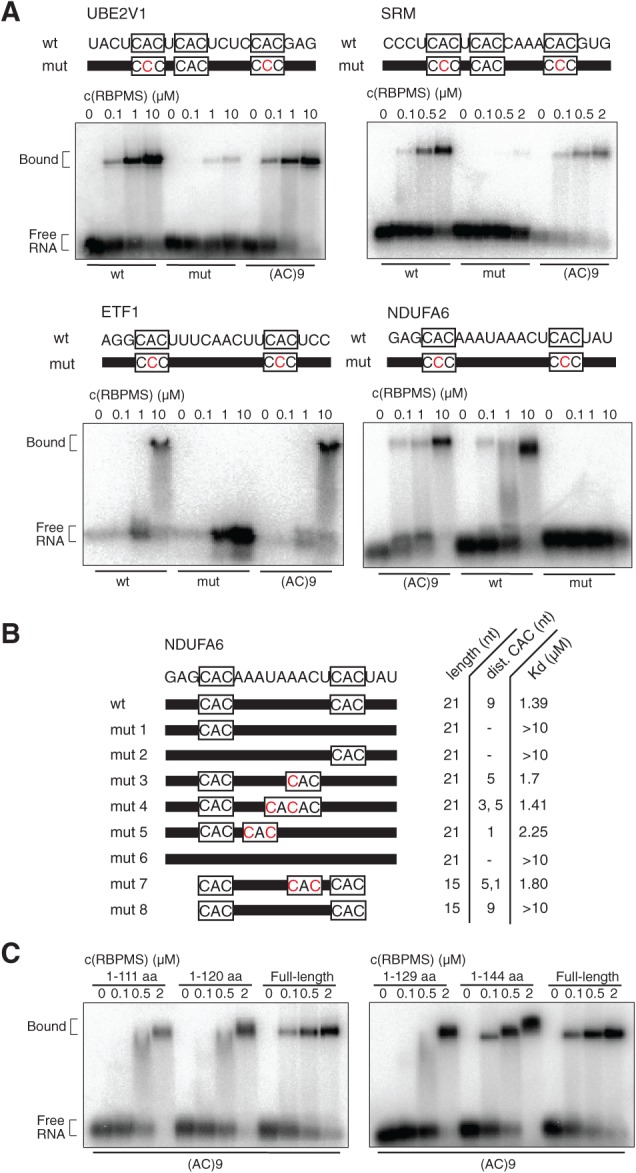
RBPMS binds PAR-CLIP targets in EMSAs. (A) 21-nt synthetic RNAs representing clusters from mRNA targets identified by PAR-CLIP (NDUFA6, UBE2V1, SRM, and ETF1) were radiolabeled (10 nM), incubated with 0–10 μM recombinant RBPMS or RBPMS2 (FLAG-HA-His6), and separated on 1% agarose gel. The RNA sequences are shown on top, with nucleotides representing the RRE boxed. Residues in red illustrate point mutations of the synthetic RNAs. (B) Dissociation constant (Kd) determination for 21- and 15-nt synthetic RNAs representing the wild-type and mutant NDUFA6 cluster. (C) Binding study of RBPMS deletion proteins (His6) to synthetic (AC)9 RNA defines additional regions contributing to RNA binding. C-terminal deletion proteins are specified; N-terminal deletion protein (amino acids 21–196) required different binding conditions and could not be directly compared (data not shown).
