FIGURE 1.
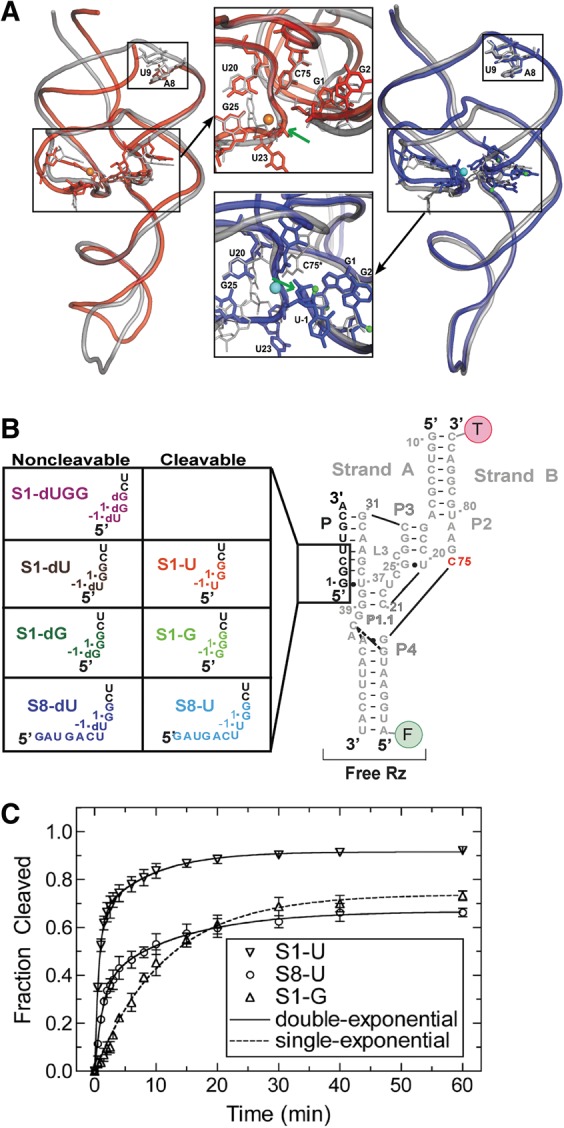
The trans-acting HDV ribozyme studied here. (A) Superpositions of trans-acting (red) (Chen et al. 2010) and cis-acting (blue) (Ke et al. 2004) precursor crystal structures on the cis-acting product structure (silver) (Ferre-D'Amare et al. 1998). Active site magnesium ions are shown in orange and cyan for the trans- and cis-acting ribozymes, respectively. The smaller black boxes on both overlays highlight the region where there is a break in the RNA backbone for the trans-acting ribozyme. The green arrows indicate the cleavage site in both insets. The U-1 nucleotide and scissile phosphate were not resolved in the trans-acting precursor structure. (B) The eight three-stranded precursor and product ribozymes used in our solution experiments. (C) Cleavage assay time courses for S1-U, S8-U, and S1-G. Cleavage assays were conducted at 25°C in 40 mM Tris-HCl, pH 7.5, 25 mM DTT, and 11 mM MgCl2. Data for S1-G were fit with a single-exponential increase function, while the data for S1-U and S8-U were best fit with a double-exponential increase function (raising the R2 as a measure of fit quality of 0.94 for single-exponentials to 0.999 for double-exponentials). Standard deviations from at least three independent assays are shown as error bars.
