Figure 1.
Identification and Characterization of the ros5 Mutants.
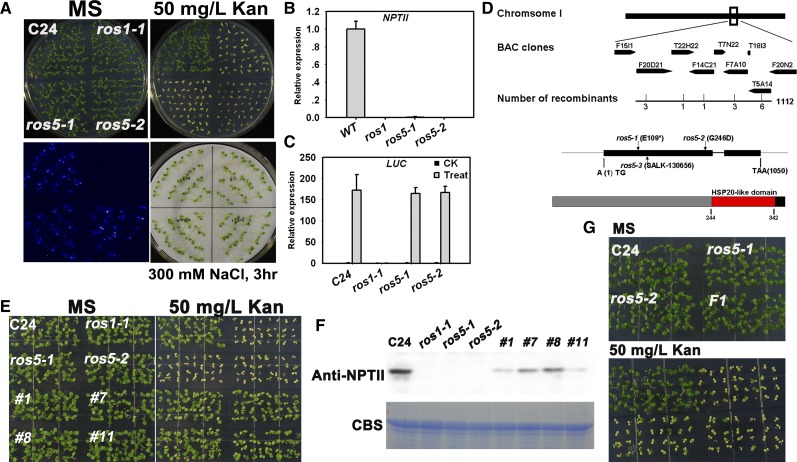
(A) The ros5 mutations lead to the silencing of 35S-NPTII but not to the silencing of the RD29A-LUC transgene. Seedlings grown on MS plates were subjected to luminescence imaging after treatment with 300 mM NaCl for 3 h. The plants were also grown on MS medium containing 50 mg/L kanamycin (Kan) and photographed after 7 d.
(B) Real-time PCR analysis of transcripts of NPTII in C24, ros1-1, ros5-1, and ros5-2. Three independent experiments were done with similar results, each with three technical replicates. One representative experiment is shown. Error bars represent se (n = 3).
(C) Real-time PCR analysis of transcripts of LUC in C24, ros1-1, ros5-1, and ros5-2 after treatment with 300 mM NaCl for 3 h. Three independent experiments were done with similar results, each with three technical replicates. One representative experiment is shown. Error bars represent se (n = 3).
(D) Diagram of the region identified by map-based cloning for ros5 mutations. In the identified region, a C-to-T mutation changes Glu to a stop codon in ros5-1 and a G-to-A mutation changes Gly to Asp in ros5-2. A T-DNA is inserted at position 20,453,546 of chromosome 1 in line SALK_130656 (ros5-3). The protein was analyzed at http://prosite.expasy.org. A conserved HSP20-like domain was found in the C-terminal region of ROS5.
(E) The kanamycin-sensitive phenotype is complemented by the ROS5 transgene in the ros5 mutants.
(F) Analysis of NPTII expression in complemented ros5 plants by protein gel blot analysis. A Coomassie blue–stained (CBS) gel was used as a loading control.
(G) Genetic analysis of the two ros5 alleles. The F1 seedlings of ros5-1 crossed with ros5-2 are sensitive on MS medium supplemented with 50 mg/L kanamycin.