Figure 1.
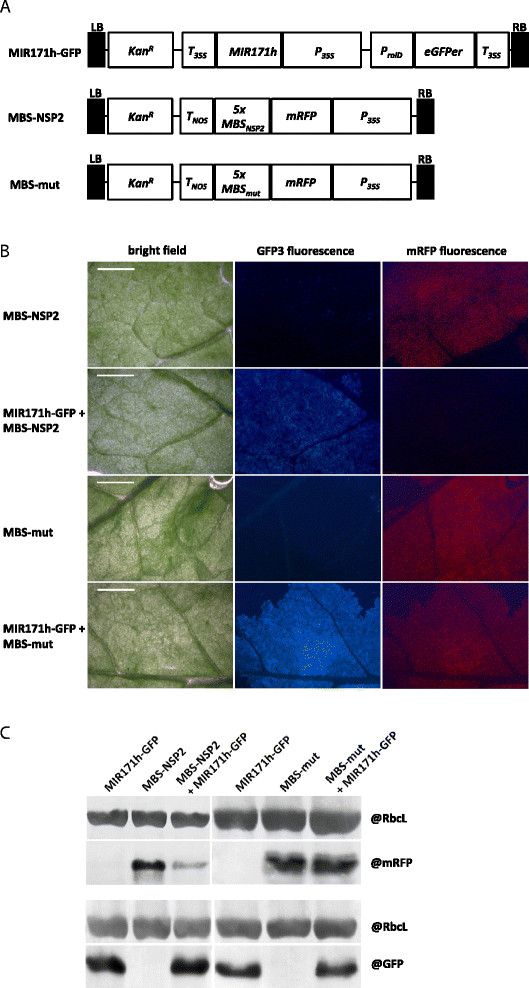
In vivo confirmation of NSP2 gene silencing by miR171h using MIR171h overexpression and mRFP sensor constructs. (A) T-DNA structure of vectors used for leave infiltration experiments. MiR171h overexpression construct (MIR171h-GFP) in pK7WG2D [[37]] and sensor constructs with either wild-type (MBS-NSP2) or mutated miR171h (MBS-mut) binding site of NSP2 cloned in pGWB455 [[38]]. LB: left boarder, KanR: kanamycin resistance gene (nptII), Tnos: nopaline synthase terminator, MIR171h: miR171h primary transcript, P35S: 35S promoter, green-fluorescent protein (GFP) cassette (pRolD–EgfpER–t35S), 5xMBSNSP2: 5 repeats of miR171h binding site sequence of NSP2, 5xMBSmut: 5 repeats of a mutated version of the miR171h binding site sequence of NSP2. (B) Co-infiltration of miR171h overexpression and mRFP sensor constructs. Nicotiana benthamiana leaves were infiltrated with the two sensor constructs MBS-NSP2 or MBS-mut. For each sensor construct, co-infiltration experiments with the MIR171h-GFP construct were carried out. Note the decreased mRFP fluorescence due to miR171h-mediated cleavage of mRFP sensor. Bright field images, GFP3 fluorescence and mRFP fluorescence are shown. Scale bar: 5 mm. (C) Western blot to prove miR171h cleavage of the miR171h binding site within the NSP2 sequence. Proteins were extracted from leaves infiltrated with MIR171h-GFP, MBS-NSP2 or MBS-mut and co-infiltration of both constructs. The upper part of the picture shows a western blot where mRFP was detected, indicating the presence of the sensor; the lower part shows a western blot with detection of GFP, indirectly indicating the presence of miR171h. On both blots, RuBisCO proteins were detected to demonstrate equal loading of the protein samples.
