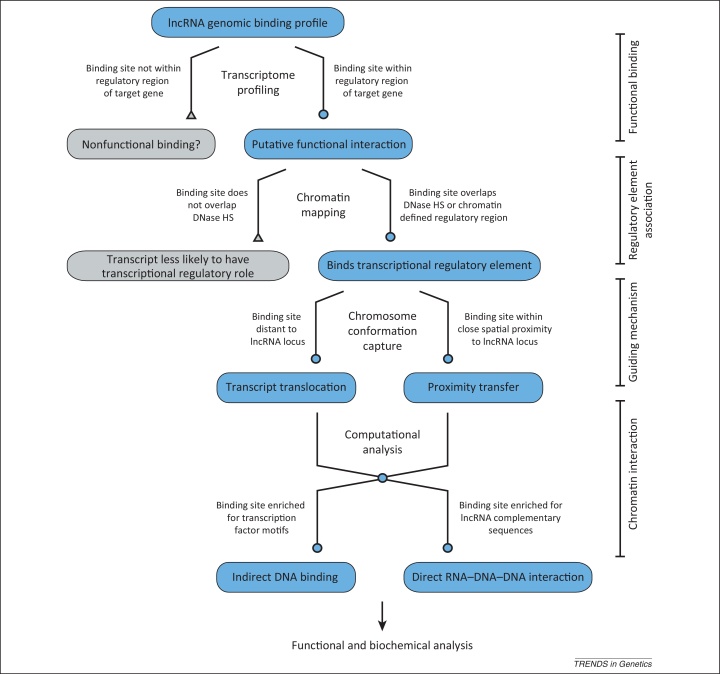
Figure 2.
Workflow diagram detailing a combined experimental and computational pipeline for investigating trans-acting long noncoding RNA (lncRNA) transcriptional regulatory functions. Likely functional lncRNA genomic binding sites are identified within the regulatory regions of target genes that are differentially expressed upon lncRNA depletion or overexpression. Gene Ontology analyses of lncRNA bound and regulated genes provide clues regarding lncRNA function. Binding sites that associate with transcriptional regulatory regions are further selected for based on DNase I hypersensitive site mapping and chromatin status. Putative functional binding locations are integrated with chromosome conformation capture-based experiments to provide insights into the mechanism of genomic targeting. Computational sequence analyses generate predictions regarding how lncRNAs interact with DNA. These criteria are used to inform the design of reporter assays and biochemical experiments aimed at understanding trans-acting lncRNA function and mode of action at candidate loci.