Table 2.
Expressed proteins identified via mass spectrometry in the embryogenic cultures of the C. papaya after 42 days in maturation treatments
| Spot a | Tratament b | Function c | Accession number d | Protein name | Organism | Peptides number e | Coverage (%) f | Score g |
Abundance
h
|
|---|---|---|---|---|---|---|---|---|---|
| Control PEG6 | |||||||||
|
Proteins of higher abundance in PEG6 treatment | |||||||||
| 017 |
Control/PEG6 |
Stress response, glycolysis |
gi|3914394 |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase |
Mesembryanthemum crystallinum |
4 |
8% |
196 |
 |
| 021 |
Control/PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
3 |
10% |
199 |
 |
| 114 |
Control/PEG6 |
Stress response |
gi|211906436 |
UDP-D-glucose pyrophosphorylase |
Gossypium hirsutum |
4 |
14% |
363 |
 |
| 128 |
PEG6 |
Stress response, Others functions |
gi|151347486 |
Methionine synthase |
Carica papaya |
3 |
5% |
70 |
 |
| 130 |
PEG6 |
Metabolic process |
gi|1743354 |
Aldehyde dehydrogenase (NAD+) |
Nicotiana tabacum |
2 |
4% |
137 |
 |
| 138 |
PEG6 |
Stress response, oxidation-redution process, Carbohydrate and energy metabolism |
gi|15222848 |
Glyceraldehyde 3-phosphate dehydrogenase |
Arabidopsis thaliana |
3 |
15% |
147 |
 |
| 139 |
PEG6 |
Protein metabolism, stress response |
gi|3914394 |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase |
Mesembryanthemum crystallinum |
2 |
3% |
68 |
 |
| 142 |
PEG6 |
Protein metabolism, stress response |
gi|21487 |
Leucine aminopeptidase |
Solanum tuberosum |
1 |
2% |
56 |
 |
| 146 |
PEG6 |
Oxidation-redution process |
gi|29501684 |
Alcohol dehydrogenase 3 |
Petunia x hybrida |
3 |
10% |
136 |
 |
| 162 |
PEG6 |
Protein metabolism, stress response |
gi|38154482 |
Molecular chaperone Hsp90-1 |
Nicotiana benthamiana |
5 |
8% |
189 |
 |
| 163 |
PEG6 |
Others functions |
gi|1707372 |
Ubiquitin-like protein |
Arabidopsis thaliana |
3 |
25% |
119 |
 |
| 165 |
PEG6 |
Unclassified |
gi|20140866 |
Full = Translationally controlled tumor protein homolog |
Cucumis melo |
2 |
13% |
83 |
 |
| 166 |
PEG6 |
Other functions |
gi|295885749 |
Cytokinin oxidase 2 |
Triticum aestivum |
1 |
1% |
46 |
 |
| 169 |
PEG6 |
Fatty acid metabolism, Carbohydrate and energy metabolism |
gi|136057 |
Full = Triosephosphate isomerase, cytosolic |
Coptis japonica |
1 |
3% |
55 |
 |
| 175 |
PEG6 |
Fatty acid metabolism |
gi|147784332 |
Hypothetical protein VITISV_041523 |
Vitis vinifera |
2 |
9% |
69 |
 |
| 184 |
PEG6 |
Metabolic process |
gi|255565327 |
Esterase D, putative |
Ricinus communis |
1 |
6% |
46 |
 |
| 188 |
PEG6 |
Carbohydrate and energy metabolism |
gi|82621108 |
Phosphoglycerate kinase-like |
Solanum tuberosum |
2 |
8% |
212 |
 |
| 194 |
PEG6 |
Oxidation-reduction process, fatty acid metabolism |
gi|2204087 |
Enoyl-ACP reductase |
Arabidopsis thaliana |
2 |
12% |
75 |
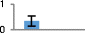 |
| 200 |
PEG6 |
Seed germination, cytoskeleton organization |
gi|32186896 |
Actin |
Gossypium hirsutum |
6 |
22% |
454 |
 |
| 215 |
PEG6 |
Oxidation-redution process, stress response |
gi|449458315 |
PREDICTED: glutamate dehydrogenase 2-like |
Cucumis sativus |
2 |
6% |
95 |
 |
| 222 |
PEG6 |
Others functions |
gi|38564733 |
Initiation factor eIF4A-15 |
Helianthus annuus |
3 |
14% |
190 |
 |
| 227 |
PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
2 |
8% |
110 |
 |
| 237 |
PEG6 |
Stress response |
gi|169661 |
S-adenosylhomocysteine hydrolase |
Petroselinum crispum |
1 |
8% |
45 |
 |
| 243 |
PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
2 |
8% |
130 |
 |
| 244 |
PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
2 |
8% |
110 |
 |
| 250 |
PEG6 |
Unclassified |
gi|462413398 |
Hypothetical protein PRUPE_ppa003850mg |
Prunus persica |
3 |
8% |
130 |
 |
|
Proteins of higher abundance in Control treatment | |||||||||
| 002 |
Control/PEG6 |
Unclassified |
gi|384249809 |
HI0933-like protein |
Coccomyxa subellipsoidea |
1 |
2% |
50 |
 |
| 011 |
Control/PEG6 |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lipase |
Carica papaya |
2 |
6% |
82 |
 |
| 012 |
Control/PEG6 |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lipase; AltName: Full = CpEST |
Carica papaya |
2 |
6% |
82 |
 |
| 018 |
Control/PEG6 |
Stress response, Carbohydrate and energy metabolism |
gi|414866626 |
TPA: hypothetical protein ZEAMMB73_999129 |
Zea mays |
2 |
6% |
64 |
 |
| 025 |
Control/PEG6 |
Oxidation-redution process |
gi|255555109 |
Flavoprotein wrbA, putative |
Ricinus communis |
1 |
11% |
112 |
 |
| 026 |
Control/PEG6 |
Other functions |
gi|162464222 |
Small GTP binding protein1 |
Zea mays |
2 |
11% |
95 |
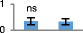 |
| 042 |
Control/PEG6 |
Carbohydrate and energy metabolism |
gi|14423688 |
Full = Enolase 1 |
Hevea brasiliensis |
2 |
8% |
62 |
 |
| 043 |
Control/PEG6 |
Stress response |
gi|2853219 |
Glutathione transferase |
Carica papaya |
5 |
22% |
330 |
 |
| 048 |
Control/PEG6 |
Others functions |
gi|89212810 |
14-3-3-like protein |
Gossypium hirsutum |
4 |
15% |
213 |
 |
| 049 |
Control/PEG6 |
Protein metabolism |
gi|399942 |
Full = Stromal 70 kDa heat shock-related protein |
Pisum sativum |
4 |
9% |
205 |
 |
| 055 |
Control/PEG6 |
Other functions |
gi|162464222 |
Small GTP binding protein1 |
Zea mays |
2 |
11% |
95 |
 |
| 054 |
Control/PEG6 |
Stress response, protein metabolism |
gi|475610277 |
Aspartic proteinase oryzasin-1 |
Aegilops tauschii |
1 |
2% |
60 |
 |
| 111 |
Control/PEG6 |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lipase |
Carica papaya |
6 |
18% |
389 |
 |
| 115 |
Control/PEG6 |
Stress response, Carbohydrate and energy metabolism |
gi|7417426 |
UDP-glucose pyrophosphorylase |
Oryza sativa Indica Group |
2 |
6% |
53 |
 |
| 315 |
Control |
Stress response, metabolic process |
gi|32527831 |
UDP-glucose pyrophosphorylase |
Populus tremula x Populus tremuloides |
3 |
9% |
122 |
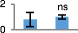 |
| 317 |
Control |
Other functions |
gi|225442221 |
Initiation factor eIF4A-15 |
Vitis vinifera |
4 |
16% |
154 |
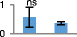 |
| 318 |
Control |
Stress response |
gi|470129411 |
PREDICTED: vicilin-like antimicrobial peptides 2-2-like |
Fragaria vesca subsp. Vesca |
1 |
1% |
49 |
 |
| 324 |
Control |
Protein folding |
gi|166770 |
Heat shock protein 83 |
Arabidopsis thaliana |
1 |
2% |
66 |
 |
| 325 |
Control |
Protein folding |
gi|20559 |
hsp70 (AA 6–651) |
Petunia x hybrid |
2 |
5% |
68 |
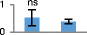 |
| 326 |
Control |
Protein folding |
gi|20559 |
hsp70 (AA 6–651) |
Petunia x hybrid |
2 |
3% |
54 |
 |
| 329 |
Control |
Stress response |
gi|226973436 |
Beta-thioglucoside glucohydrolase |
Carica papaya |
3 |
9% |
165 |
 |
| 333 |
Control |
Oxidation-redution process |
gi|295367043 |
Cinnamoyl alcohol dehydrogenase |
Bambusa multiplex |
5 |
23% |
48 |
 |
| 335 |
Control |
Carbohydrate and enregy metabolism |
gi|359483362 |
PREDICTED: lactoylglutathione lyase |
Vitis vinifera |
3 |
11% |
146 |
 |
| 337 |
Control |
Protein folding |
gi|62433284 |
BiP |
Glycine Max |
2 |
5% |
75 |
 |
| 339 |
Control |
Stress response |
gi|226973436 |
Beta-thioglucoside glucohydrolase |
Prunus pérsica |
3 |
6% |
126 |
 |
| 340 |
Control |
Stress response |
gi|226973436 |
Beta-thioglucoside glucohydrolase |
Carica papaya |
3 |
6% |
108 |
 |
| 343 |
Control |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lípase |
Carica papaya |
2 |
6% |
163 |
 |
| 345 |
Control |
Carbohydrate and energy metabolism |
gi|356562473 |
PREDICTED: xylose isomerase-like |
Glycine Max |
3 |
9% |
56 |
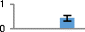 |
| 347 |
Control |
Other functions |
gi|357440579 |
Pentatricopeptide repeat-containing protein |
Medicago truncatula |
7 |
12% |
57 |
 |
|
Proteins expressed similarly in both treatments | |||||||||
| 015 |
Control/PEG6 |
Stress response, metabolic process |
gi|211906436 |
UDP-D-glucose pyrophosphorylase |
Gossypium hirsutum |
5 |
15% |
213 |
 |
| 024 |
Control/PEG6 |
Other functions |
gi|148912162 |
Cytosolic ascorbate peroxidase 1 |
Gossypium hirsutum |
3 |
20% |
221 |
 |
| 027 |
Control/PEG6 |
Oxidation-redution process |
gi|449464176 |
PREDICTED: proteasome subunit beta type-3-A-like |
Cucumis sativus |
2 |
15% |
175 |
 |
| 028 |
Control/PEG6 |
Metabolic process |
gi|5669924 |
Soluble inorganic pyrophosphatase |
Populus tremula x Populus tremuloides |
1 |
7% |
37 |
 |
| 032 |
Control/PEG6 |
Stress response |
gi|226973430 |
Beta-thioglucoside glucohydrolase |
Carica papaya |
5 |
13% |
278 |
 |
| 044 |
Control/PEG6 |
Unclassified |
gi|412993224 |
ORF73 |
Bathycoccus prasinos |
1 |
0% |
36 |
 |
| 045 |
Control/PEG6 |
Other functions |
gi|270313547 |
S-adenosylmethionine decarboxylase |
Olea europaea |
1 |
2% |
47 |
 |
| 051 |
Control/PEG6 |
Protein metabolism, Stress response |
gi|233955399 |
Calreticulin |
Carica papaya |
6 |
22% |
323 |
 |
| 052 |
Control/PEG6 |
Oxidation-redution process |
gi|193290660 |
Putative ketol-acid reductoisomerase |
Capsicum annuum |
3 |
10% |
116 |
 |
| 056 |
Control/PEG6 |
Unclassified |
gi|384249809 |
HI0933-like protein |
Coccomyxa subellipsoidea |
1 |
2% |
58 |
 |
| 060 |
Control/PEG6 |
Oxidation-redution process |
gi|449520553 |
PREDICTED: superoxide dismutase [Mn], mitochondrial-like isoform 1 |
Cucumis sativus |
2 |
13% |
204 |
 |
| 061 |
Control/PEG6 |
Oxidation-redution process |
gi|33521626 |
Mn-superoxide dismutase |
Lotus japonicus |
1 |
8% |
65 |
 |
| 062 |
Control/PEG6 |
Unclassified |
gi|384253982 |
Hypothetical protein COCSUDRAFT_39121 |
Coccomyxa subellipsoidea C-169 |
1 |
1% |
44 |
 |
| 067 |
Control/PEG6 |
Unclassified |
gi|460412635 |
PREDICTED: 20 kDa chaperonin, chloroplastic-like |
Solanum lycopersicum |
3 |
12% |
221 |
 |
| 069 |
Control/PEG6 |
Stress response, signaling |
gi|2853219 |
Glutathione transferase |
Carica papaya |
2 |
10% |
41 |
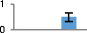 |
| 087 |
Control/PEG6 |
Others functions |
gi|55375985 |
14-3-3 family protein |
Malus x domestica |
6 |
33% |
504 |
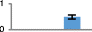 |
| 095 |
Control/PEG6 |
Stress response |
gi|357514981 |
Annexin-like protein |
Medicago truncatula |
2 |
5% |
57 |
 |
| 099 |
Control/PEG6 |
Strress response |
gi|356531939 |
PREDICTED: putative lactoylglutathione lyase-like |
Glycine Max |
3 |
9% |
152 |
 |
| 126 |
Control/PEG6 |
Metabolic process |
gi|5725356 |
Alpha-D-xylosidase |
Tropaeolum majus |
1 |
1% |
37 |
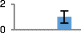 |
| 003 |
Control/PEG6 |
Unclassified |
gi|357498189 |
DNA repair and recombination protein PIF1 |
Medicago truncatula |
1 |
2% |
40 |
 |
| 007 | Control/PEG6 | Oxidation-redution process | gi|12322163 | Dormancy related protein, putative | Arabidopsis thaliana | 1 | 3% | 47 |  |
PEG: polyethylene glycol.
aSpot numbers correspond to the numbers indicated in Figure 3.
bMaturation treatments: Control - without PEG; and PEG6 - with 6% PEG.
cFunctional protein classification using Blast2go (http://www.blast2go.com).
dAccession number in the NCBI protein database.
eNumber of unique peptide sequences identified by MASCOT.
fPercentage of predicted protein sequence covered by MASCOT.
gProbability-based MOWSE score from MASCOT software for the hit.
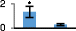
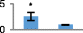
hProtein relative abundance according to the individual spot (%) volume determined using Image Master Platinum v.7 software (GE Healthcare). Change in abundance levels by t-test, *(p ≤ 0.05), **(p ≤ 0.01), ns (not significant).
