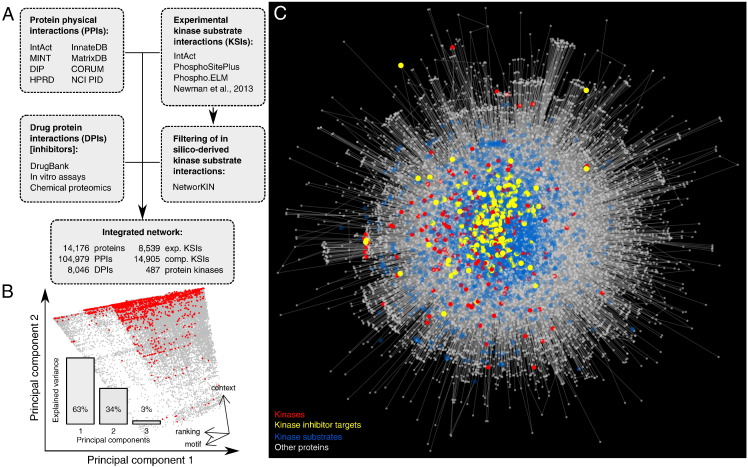
Fig. 1.
Construction of a protein kinase-relevant interaction networks. (A) Multiple sources of interaction data were integrated combining protein–protein interactions (PPIs), kinase to substrate interactions (KSIs) and inhibitory drug to kinase interactions (DPIs). (B) To complement experimentally determined KSIs, we included filtered in silico KSIs from NetworKIN. Principal component analysis of the three scores provided by NetworKIN and intersection with experimental KSIs (red) identified a well-defined score range to filter NetworKIN KSIs. Variance explained by the principal components (bottom left) and orientation of the original coordinates in the space spanned by the first 2 principal components (bottom right). (C) The resulting reference network.