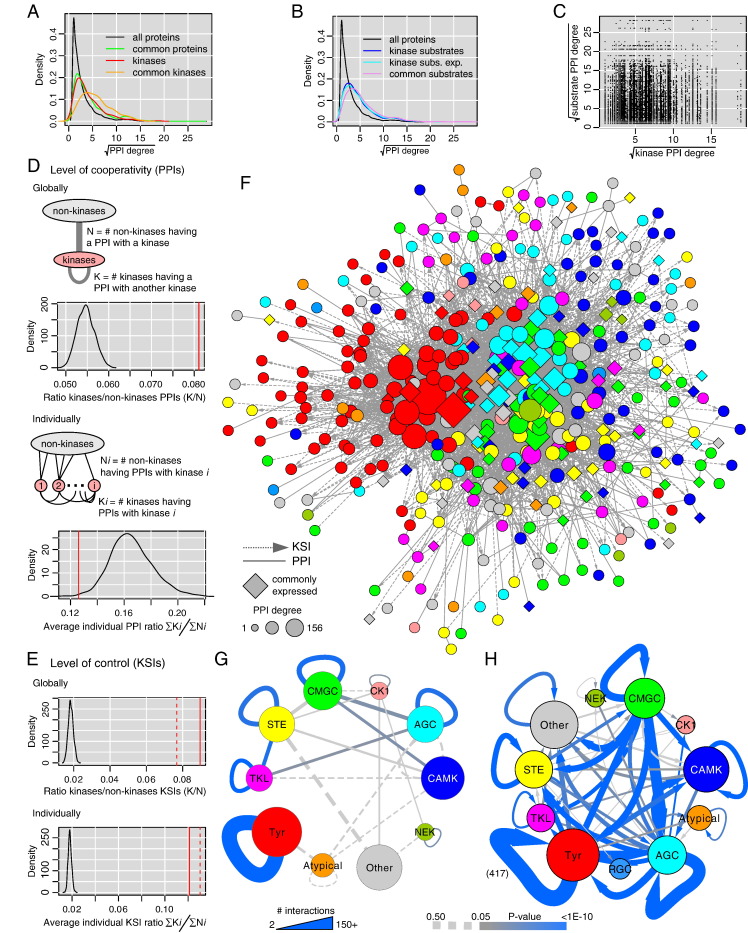
Fig. 3.
Connectivity of kinases. (A) Kinases tend to have more PPIs with other proteins than average and this trend is further augmented for commonly expressed kinases that are identified in almost all cell types. (B) Similar observation for kinase substrates. Substrates known from experimental data only have the same bias (cyan). (C) The number of PPIs of a kinase is not correlated with the one of its substrates. (D) Cooperation with other kinases. Globally, the pooled kinase PPIs touch a set of human proteins including 8% of kinases (red line), which is much more than expected by chance (black null distribution). On the contrary, the average ratio of PPIs linking an individual kinase to other kinases versus non-kinases is significantly less than expected by chance. (E) Regulation (control) of other proteins through phosphorylation. Both globally and individually, kinases control other kinases significantly more than expected by chance. (F) Kinase–kinase network linking all the kinases for which we could obtain interaction data (487). Colors associated to each kinase group are given in panels G and H. (G) Analysis of cooperation (PPIs) within and across kinase groups. Since a few interconnections only were significant (P-value < 0.05) we featured edges with at least 3 PPIs and P-value < 0.5 as dashed lines. (H) Same analysis for regulation (KSIs). One self-connection (Tyr to Tyr) was supported by such a large number of KSIs (417) that it was outside the line width scale used to plot the network.