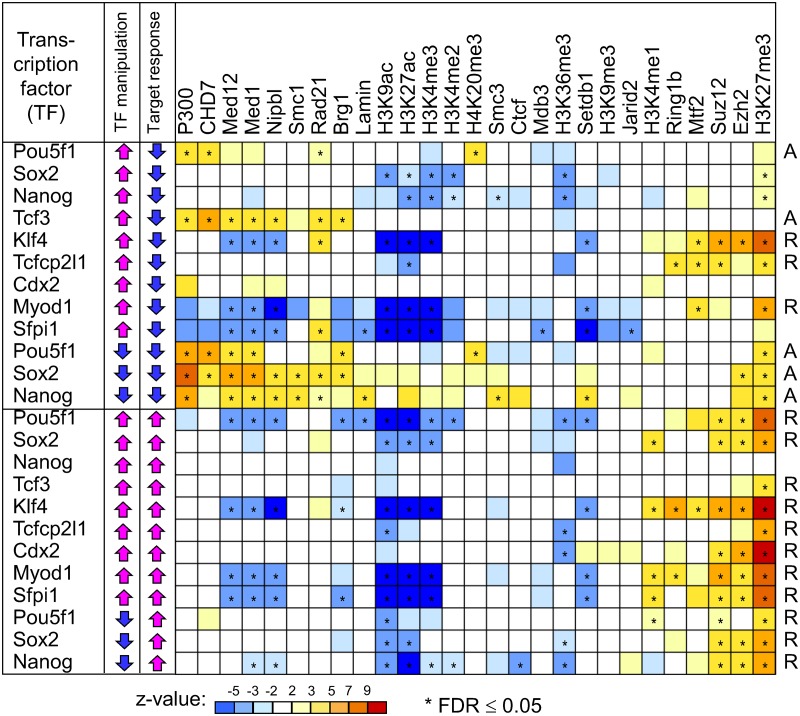
FIG. 2.
Correlation analysis of the response of target genes to manipulations of transcription factors (TFs) and presence of chromatin modifications and cofactors at TF binding sites. Correlation patterns reveal two kinds of functional TF binding sites: (1) active sites (marked “A” at right) are bound by P300, CHD7, mediator (Med1, Med12), cohesin (Smc1, Nipbl, Rad21), and SWI/SNF (Brg1), and (2) repressed sites (marked “R” at right) carry repressive histone marks H3K27me3 and are bound by Polycomb TFs (Ezh2, Suz12, Mtf2, and Ring1b). Color corresponds to the average z-value from seven types of analysis, including three algorithms for estimating factor abundance, rank correlation, and multivariate regression; statistical significance is evaluated using false discovery rate (FDR) estimated from permutations of gene expression change. Effects marked as significant satisfy condition FDR ≤ 0.05 in all 7 methods of analysis.