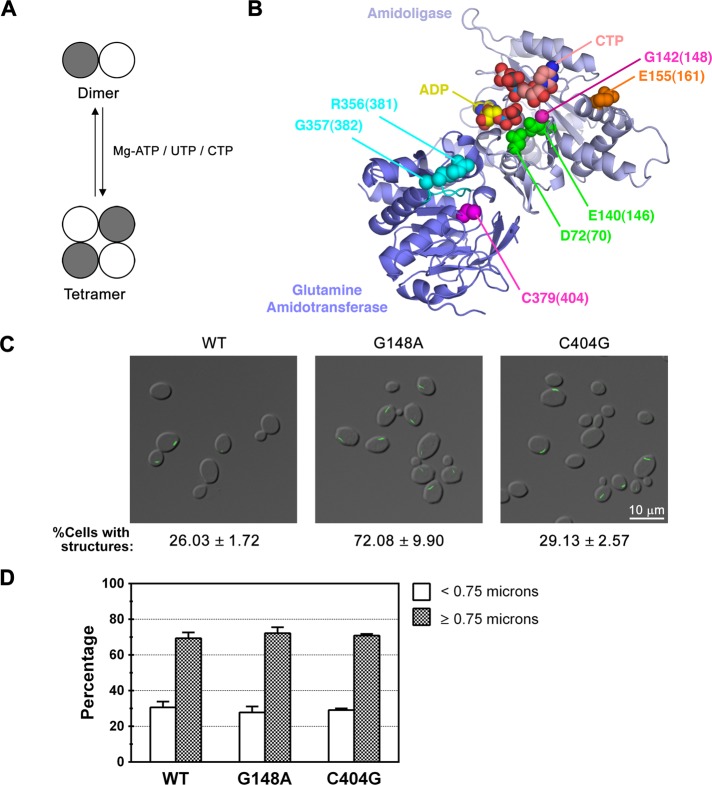
FIGURE 2.
Effect of disrupting UTP-mediated tetramerization or the active site of CTP synthase on filament formation. (A) The transition of CTP synthase from an inactive dimer to an active tetramer is regulated by ATP, UTP, and CTP binding. (B) Crystal structure of E. coli CTP synthase (PDB:2AD5; Endrizzi et al., 2005), highlighting residues involved in Mg2+ATP binding/tetramerization (D72, E140; green), UTP binding/tetramerization (G142; magenta), CTP binding/tetramerization (E155; orange), GTP binding (R356/G357; cyan), and catalysis (C379; magenta). The amidoligase domain (residues 1–266) and glutamine amidotransferase domain (residues 287–544)/interdomain linker (267–286) are colored light and dark blue, respectively, and the L11 lid is colored cyan. The bound ADP and CTP molecules in the structure are colored yellow and pink, respectively. The numbers in parentheses represent the corresponding amino acids in Ura7p. A multiple sequence alignment noting the targeted residues is also provided for reference (Supplemental Figure S2). (C) Representative images of yeast strains expressing wild-type (WT), catalytic mutant, or tetramerization mutant Ura7p-GFP. The average percentage and SEM of cells with Ura7p-GFP structures are indicated below each image. (D) Percentages of foci (<0.75 μm) and filaments (≥0.75 μm) for Ura7p-GFP structures are graphed for WT Ura7p-GFP and each mutant for comparison. Protein levels were uncorrelated with the effects on filament frequency or length (Supplemental Table S1 and Supplemental Figure S1).