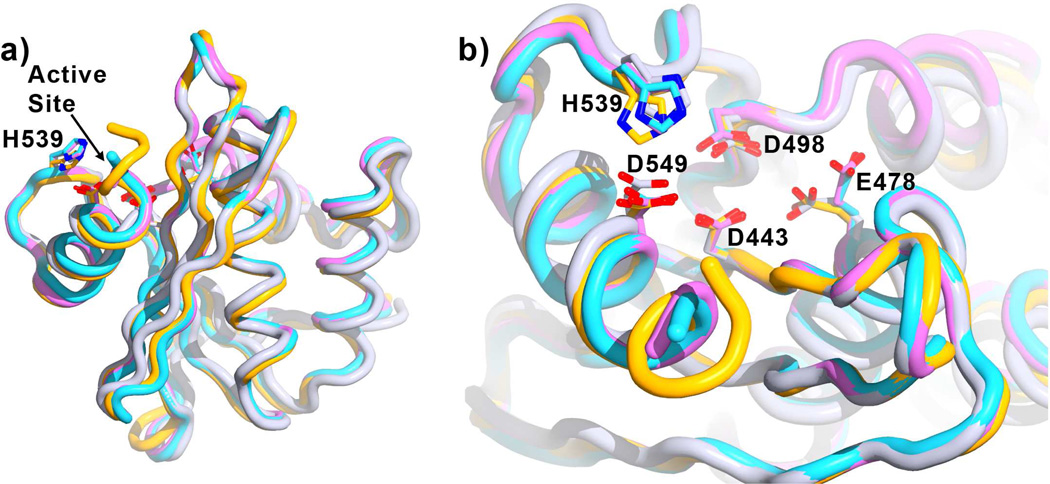
Figure 4. Effect of F3284-8495 and cations on RNase H conformation.
Shown is the superposition of the current structure (orange, inhibitor not shown) with the RNase H domain of several RT structures without inhibitors bound at the RNase H active site (referenced here by Protein Data Bank accession numbers): 3DLK53 (cyan), with no ligands bound; 4G1Q54 (violet), with Mg2+ bound at the Cation A position; and 2BE255 (gray), with Mn2+ bound at the Cation B position. Superposition is based on main-chain atoms for residues 441 to 448 of the RNase H domain. a) A view of the entire RNase H domain. The overall conformation of the RNase H domain is highly similar whether or not ligands are bound. b) A closer view of the active site, rotated toward the viewer by about 60 degrees. Glu478 tends to point away from the active site if there is no Cation B to coordinate (3DLK and 4G1Q). His539 may adjust its position and side-chain conformation to interact with a ligand such as F3284-8495 at the active site.