Figure 2.
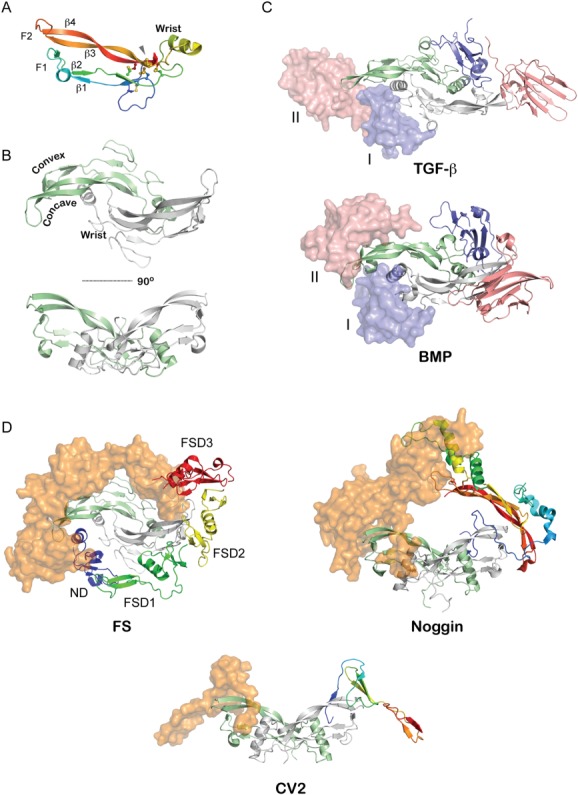
Structure of TGF-β ligands and their associated complexes. A: Representative structure of a TGF-β ligand monomer (Myostatin from PDB 3HH2).37 Intramolecular disulfide bonds are shown as sticks. Labels indicate various β-stands in the ligand as well as identify the finger-wrist-finger architecture of the ligands. B: Structure of the mature TGF-β ligand homodimer (PDB 3HH2) with one monomer colored in light green and another in gray.37 The monomers are linked via an intermolecular disulfide bond. This architecture exposes extended convex and concave surfaces on the protein, which have a strong hydrophobic character and define the ability of the protein to interact with its cognate receptors. C: Structure of TGF-β ligands bound to their Type I and Type II receptors (PDB 2PJY).93 The ligand is represented in ribbon (green, gray) with the receptors represented in both surface and ribbon representation (pink, blue). The receptors for the TGF-β subclass (top) come into contact during binding since the Type I receptors binds more towards the fingertips of the ligand while those for the BMP subclass (bottom) do not, where the Type I receptor binds more significantly to the exposed convex epitope of BMP ligands (PDB 2GOO).94 D: Representative structures of various BMP-antagonist complexes. Ligands are indicated by ribbon diagrams (green, gray), while antagonists are shown as rainbow colored ribbons (one half) and orange surface representations. (left) FS-Myostatin complex (PDB 3HH2).37 Labels indicate various domains of FS. (middle) Noggin-BMP7 complex (PDB 1M4U).34 (right) CV2-BMP2 complex (PDB 3BK3).39 A single VWC domain was solved in complex from the larger multidomain CV2 protein.
