Abstract
Engineering fluorescent proteins (FPs) to emit light at longer wavelengths is a significant focus in the development of the next generation of fluorescent biomarkers, as far-red light penetrates tissue with minimal absorption, allowing better imaging inside of biological hosts. Structure-guided design and directed evolution have led to the discovery of red FPs with significant bathochromic shifts to their emission. Here, we present the crystal structure of one of the most bathochromically shifted FPs reported to date, AQ143, a nine-point mutant of aeCP597, a chromoprotein from Actinia equina. The 2.19 Å resolution structure reveals several important chromophore interactions that contribute to the protein's far-red emission and shows dual occupancy of the green and red chromophores.
Keywords: red fluorescent protein; near-infrared, bathochromic shift; chromoprotein
Introduction
Fluorescent proteins (FPs) that emit light in the near-infrared (NIR) window (∼650–900 nm) are in demand as biological imaging agents. The NIR window is a local minimum at which light penetrates tissue with minimal absorption from biological molecules such as melanin, hemoglobin, and water.1 FPs natively do not emit light in the NIR; the longest maximum intensity emission wavelength (λem) reported to date for a native red FP (RFP) is 613 nm, found in NvFP-7R from Nematostella vectensis.2 FPs with significant bathochromic shifts to λem have been produced with both rational design and directed evolution but these molecules tend to have low quantum yields, poor brightness, and other characteristics that compromise their utility.3–8 Many FP engineering strategies, including those that have induced bathochromic shifts in the λem, have relied on atomic-resolution structural data to guide intuition-based design, motivating continued efforts to obtain additional structural information for far-red FPs. AQ143, which was engineered from aeCP597, a chromoprotein from Actinia equine,9 is one of only seven known FPs of the Aequorea victoria FP-like superfamily that exhibit a peak emission wavelength of at least 650 nm. The other six proteins (Neptune,10 eqFP650,7 TagRFP657,11 mCardinal,12 eqFP670,7 and TagRFP6758) are all variants of eqFP578, a native RFP from Entacmaea quadricolor.13 There are known structures for five of these proteins (Neptune: 3IP2, eqFP650: 4EDO, mCardinal: 4OQW, eqFP670: 4EDS, TagRFP675: 4KGF), but as they are all derived from the same ancestral protein, there is limited sequence diversity among these structures. Here we report the 2.19 Å crystal structure of AQ143, which is derived from a more distantly related protein, aeCP597 (∼60% sequence identity to eqFP578 and its variants). AQ143 has a novel chromophore environment (defined as all internal-facing residues within 5 Å of the chromophore), which shares no more than 70% (16 of 23 positions) sequence identity with any other FP. Glu41 plays an important role in red-shifting AQ143's emission spectrum and is not seen in any other FP. The reported structure also provides evidence in support of recently reported red-shifting chromophore interactions.6,8,14
Results and Discussion
The asymmetric unit contains eight protein molecules, which align with an all-atom r.m.s.d. of 0.27–0.74 Å with differences between molecules concentrated mostly in the loop regions and in the C- and N-terminal tails. The chromophore region also varies somewhat between molecules and shows weak electron density around the phenolate side chain, which could be attributed to mobility in the phenolate side chain and co-occupancy of two different chromophores—green and red.
Oligomerization
AQ143 is a native tetramer, which is clear in the crystal packing. The asymmetric unit, however, contains eight monomers, or two such tetrameric assemblies with the C-terminal tail of each monomer involved in making intertetramer contacts. To verify the oligomerization state of AQ143, we ran both size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC). SEC analysis indicates that AQ143 behaves as a tetramer, but that it has slight octomeric properties, while AUC confirms that the protein is predominantly tetrameric (Supporting Information Figs. S1 and S2). As oligomerization is an important consideration in the engineering of RFPs, all of which are natively tetrameric,15 we compared the AB and AC interfaces of AQ143 with those of four other native RFPs (Supporting Information Table S1) using the PISA server of the European Bioinformatics Institute16 and report average buried surface area and average ΔiG (the solvation free energy gain on formation of the interface). The AC interface is known to be the tighter of the two interfaces,17 which is consistent with the AC interface having more negative ΔiGs. Interestingly, although AQ143 showed a similar amount of buried AB interface surface area, the ΔiG for AQ143's AB interface is very high, indicating a large amount of hydrophobic residues at this interface. The AC interface is more difficult to compare as the amount of buried surface area varies widely, although this is in part due to the lack of crystallographic density at the C-terminal tails (which participate intimately in this interface) in many of these structures. DsRed, the first successfully monomerized RFP17 shows the lowest ΔiG for its AC interface, possibly indicating that future monomerization efforts of AQ143 may be more difficult.
Green and red chromophores
Many engineered far-red FPs exhibit slow or incomplete maturation to the red chromophore,4,11,18 and it has recently been shown that maturation to the green and red chromophores in DsRed-type FPs occurs via a branched pathway (i.e., the two forms of the chromophore are separate endpoints in chromophore maturation; the green is not an intermediate in the maturation to the red chromophore as had been previously proposed).19 AQ143 is a DsRed-type FP with a chromophore composed of a methionine/tyrosine/glycine triad that matures to both a green and a red chromophore (Fig. 1), as evidenced by its absorbance, excitation, and emission spectra (Supporting Information Figs. S3–S5). To calculate the percentage of chromophores that mature to the green and to the red, we determined the extinction coefficients of the two species by the dynamic difference method. In this procedure, AQ143 was pH-adjusted to alkaline conditions, in which the green and red chromophores denature at different rates and their respective contributions to the 450 nm alkali-denatured absorbance peak can be determined (Supporting Information Figs. S6–S8 and Methods).5 We calculated the extinction coefficient to be 58,000 ± 11,000 M−1 cm−1 for the red chromophore and 47,000 ± 5,000 M−1 cm−1 for the green chromophore. From these data, we estimated the percentages in the fully mature protein to be 33 ± 6% for the red and 67 ± 6% for the green chromophore. Measurements of the protein in the crystal condition suggested that this fraction did not change on crystallization. Corroborating the spectroscopic evidence, we observed that the refined electron density map of AQ143 shows a mixture of chromophores containing both the oxidized N-acylimine (red) and the unoxodized N-acylamine (green) at the N-terminal residue of the chromophoric triad. The estimated occupancy of the red and green chromophores averaged across all eight monomers in the asymmetric unit is 24 ± 9% and 76%, respectively. Thus the spectroscopic calculations of chromophore occupancy in the crystal condition are consistent with the crystallographic refinement.
Figure 1.
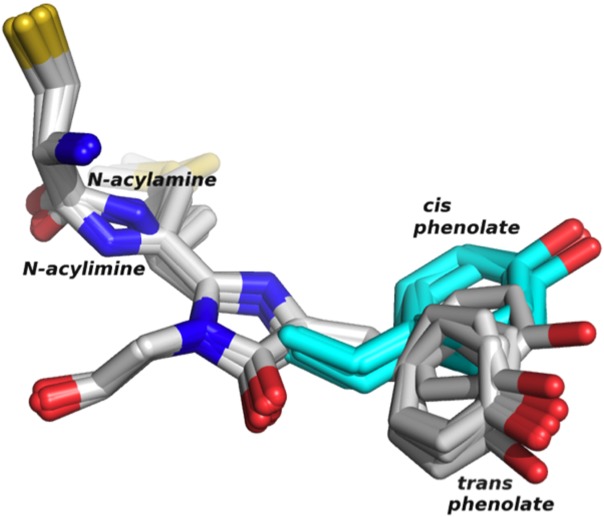
Alignment of the chromophores and C-terminal cysteine from each of the eight monomers in the asymmetric unit. The modeled cis phenolate is shown in turquoise. The N-acylamine and N-acylimine are present in the green and red chromophores, respectively.
Cis versus trans phenolate
The phenolate side chain of the chromophore (the phenolate group) in DsRed-type FPs and related chromoproteins can occupy either a cis or a trans conformation, indicating its proximity to the N1 nitrogen of the imidazolinone ring of the chromophore. For many RFPs, a trans to cis isomerization of this phenolate moiety, which is sometimes pH-inducible,20,21 has been implicated in fluorescence. In nonfluorescent chromoproteins, for instance, the chromophore is found in the trans conformation, and mutations to these chromoproteins that stabilize the cis conformation have created FPs such as HcRed and AQ143. In engineering AQ143 from the chromoprotein aeCP597, Cys143Ser was reported to be responsible for inducing weak fluorescence,9 as the mutation to serine stabilizes the cis chromophore by providing a hydrogen bond to the hydroxyl oxygen of the phenolate side chain. In the referenced work, fluorescence was improved by removing a serine hydrogen bond to the hydroxyl of the trans phenolate with a Ser158Ala mutation, further stabilizing the cis over the trans chromophore. By inducing fluorescence in an otherwise nonfluorescent chromoprotein, these mutations seem to imply that the cis chromophore represents the fluorescent moiety in AQ143.
Indeed, the refined structure shows good electron density for all parts of the chromophore with the exception of the phenolate side chain, which we modeled in the trans configuration. However, the difference map shows that the modeled phenolate is not a perfect fit, as the electron density is not sufficient to describe a chromophore that is solely found in the modeled trans configuration, while residual density appears in the position we expect that the cis phenolate would occupy. The refined electron density is such that we expect there is a co-occupancy in the crystal of two or more chromophore orientations and also possibly that the phenolate is mobile in one or both of these chromophore species. This would be consistent with a cis–trans isomerization of the chromophore on fluorescence excitation, as has been seen in other FPs.5,22,23 The lack of clear electron density for the phenolate moiety implies that the fluorescence-inducing mutations in AQ143 may have had their predicted effects, namely in destabilizing the native trans chromophore, and allowing for the phenolate to occupy the cis conformation. Given the ambiguity associated with the chromophore orientation and the lack of clear density for the cis conformation, we elected to model-build the cis phenolate postrefinement (Fig. 1). The modeled position of the cis phenolate accommodates a hydrogen bond between the hydroxyl of the fluorescence-inducing Cys143Ser mutation and the phenolate oxygen, supporting the hypothesis that this interaction is linked to the induction of fluorescence in AQ143 (Fig. 2). A second water-mediated hydrogen bond to the phenolate oxygen appears to further stabilize the cis conformation.
Figure 2.
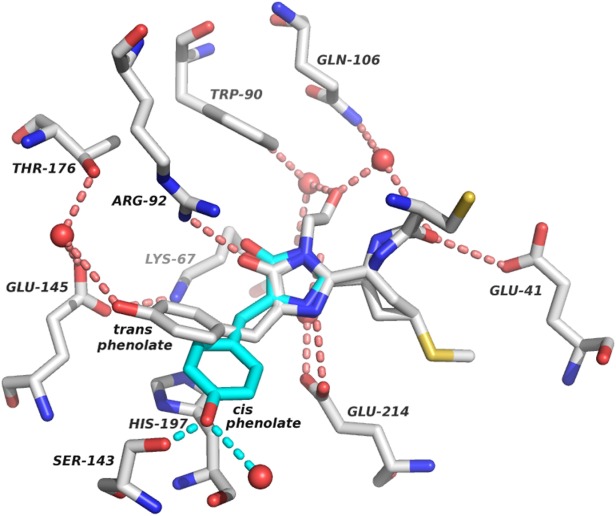
Chromophore contacts in AQ143. Residues that directly interact with the chromophore or help to co-ordinate structural waters (red spheres) are shown along with the immediate hydrogen-bonding network. A representative chromophore was chosen (chain E) to illustrate the contacts. Hydrogen bonds (dotted lines) are shown for interactions with the chromophore. The modeled cis conformation is shown in turquoise, along with two putative hydrogen bonds to its hydroxyl group. Two hydrogen bonds to the acylimine oxygen from Glu41 and a co-ordinated water can be seen in the right of the figure.
Interestingly, neither the trans nor the modeled cis conformations of the chromophore are coplanar with the imidazoline ring. This noncoplanarity is relatively uncommon in FPs and has been proposed to be responsible for low quantum yields.10 AQ143 indeed has a very low quantum yield (0.04),9 and improving the coplanarity of the two chromophore rings may represent an opportunity to further improve its fluorescence.
Mechanisms of bathochromic shift
AQ143 exhibits a number of red-shifting chromophore interactions that have been well documented in the literature.8 A network of direct and water-mediated hydrogen bonds has been proposed to lower the energy of the photoexcited state of the chromophore's conjugated π-electron system, resulting in bathochromic shifts to λem.10,24 In AQ143, three hydrogen bonds to the chromophore are good candidates to provide such stabilization including two hydrogen bonds to the acylimine oxygen, as well as one to the phenolate oxygen (Fig. 2).
Glu41 and a water molecule co-ordinated by Gln106 and the chromophore's C-terminal acyl oxygen both form hydrogen bonds to the chromophore N-acylimine (Fig. 2). To our knowledge, the only other FPs known to have two hydrogen bonds to the acylimine oxygen are CjBlue, the furthest red-shifted chromoprotein, and TagRFP675, the furthest red-shifted FP, although in TagRFP675, the hydrogen bond donor at the position equivalent to Glu41 in AQ143 is a glutamine.8,14 mPlum, the furthest red-shifted monomeric FP,3 has a similar hydrogen bonding interaction between Glu16 and the chromophore N-acylimine, but is lacking a co-ordinated water molecule to provide the second hydrogen bond. The importance of hydrogen bonds to the N-acylimine was shown in mPlum variants, in which Glu16 is mutated to other residues including proline and glutamine, causing significant hypsochromic shifts to λem.18,25,26
Additionally, flexibility in the hydrogen bonding network to the phenolate oxygen of the chromophore, particularly via water-mediated hydrogen bonds, has been proposed to be responsible for extended stokes' shifts and significant bathochromic shifts to fluorescence emission.8 The modeled cis chromophore, which we believe to be the fluorescent moiety, can accommodate two hydrogen bonds to the phenolate oxygen from the hydroxyl of Ser143 and a structural water molecule (Fig. 2). The trans chromophore, despite the mutation away from Ser158, makes a hydrogen bond contact with a structural water molecule stabilized by Glu145 and Thr176, although the effect of this interaction is less clear as the trans chromophore is not thought to be fluorescent.
Finally, many red-shifted FPs have been described that exhibit π-stacking interactions with the phenolate group of the chromophore.6,27,28 Histidine and tyrosine have both been reported at positions analogous to His197 in AQ143 with histidine present in eqFP578, RFP639, and mRuby13,29,30, and tyrosine present in mRojoA, TagRFP657, and mGrape36,10,11. In engineering mRojoA, a tyrosine π-stacking interaction with the cis phenolate was explicitly designed into the protein which resulted in a 7 nm red-shift6. In AQ143, His197 appears to form a π-stacking interaction with the trans phenolate (Fig. 2), which we presume to be the nonfluorescent entity. Interestingly, in mRuby and eqFP578, the histidine also π-stacks with the trans phenolate, whereas in the further red-shifted RFP639, the π-stack occurs with the cis phenolate. This implies that there may be room to further stabilize the photo-excited state of the cis phenolate of AQ143 and red-shift its emission by optimizing the π-stacking interaction with the cis chromophore.
Conclusions
AQ143 is one of the furthest red-emitting FPs of the GFP family, and the structure reported in this study helps elucidate some of the features underlying its far-red emission. A recently reported FP, TagRFP675, shares many of the same chromophore interactions responsible for AQ143's bathochromic shift.8
Materials and Methods
Protein expression and purification
A synthetic gene construct encoding an N-terminal polyhistidine tagged AQ143 (GenBank KF479351) was assembled in vitro, expressed in Escherichia coli BL21(DE3) cells, purified, and crystallized. Cultures were grown at 37°C to an optical density of ∼0.6 in LB, induced, then allowed to express protein at 20°C for 24 h. Protein was purified via His-tag affinity chromatography, run over a size exclusion column to remove trace contaminants and move the protein into storage buffer (1× phosphate buffered saline (PBS) pH 7.4), and finally concentrated to 18 mg/ml.
Crystallization, data collection, and structure determination
Rectangular plate crystals grew in 7 days by the sitting-drop vapor diffusion method in 100 mM Tris pH 7.0 with 50 mM lithium sulfate and 20% w/v PEG 3350. Crystals were flash frozen in 2-methyl-2,4-pentanediol and shipped to beamline 12-2 at the Stanford Synchrotron Radiation Lightsource, where a 2.19 Å data set was collected. Phases were obtained through molecular replacement using the crystal structure of the FP asFP595 (PDB ID 1A50).
Following molecular replacement, model building and refinement were run with COOT and PHENIX.31,32 non crystallographic symmetry (NCS) restraints were applied to early refinement steps and removed at the final stages of refinement. Translation/libration/screw (TLS) parameters were used throughout. The chromophore was initially left out of the refinement and added at a later stage when clear density became evident for it. First, the chromophore was added without the phenolate side chain, as little density appeared for this group. Subsequently, as density became clearer, a trans chromophore was added. The final modeled chromophore has a trans phenolate ring, an imidazoline heterocyclic ring, and dual occupancy of a green N-acylamine and a red N-acylimine. Co-ordinates were deposited in the Protein Data Bank with the code 4OHS. Data collection and refinement statistics are listed in Table I.
Table I.
X-Ray Data Reduction and Crystallographic Refinement Statistics
| (A) X-ray data reduction statistics | |
| Space group | P1 |
| Unit cell dimensions (a, b, c) | 51.0, 68.1, 132.8 Å |
| Resolution | 39.1–2.19 Å |
| (last shell) | 2.31–2.19 Å |
| Total measurements (last shell) | 281,018 (30,290) |
| Number of unique reflections (last shell) | 72,946 (8028) |
| Wavelength | |
| R-merge (last shell) | 0.072 (0.749) |
| I/σ(I) (last shell) | 11.9 (1.7) |
| Completeness (last shell) | 0.861 (0.648) |
| Multiplicity (last shell) | 3.9 (3.8) |
| (B) Crystallographic refinement statistics | |
| Resolution | 131.1–2.19 Å |
| (last shell) | 2.22–2.19 Å |
| No. of reflections (working set) | 69,234 |
| No. of reflections (test set) | 3647 |
| R-factor (last shell) | 0.190 (0.315) |
| R-free (last shell) | 0.221 (0.338) |
| No. of amino acid residues | 1,770 |
| No. of atoms | 14,508 |
| No. of solvent molecules | 355 |
| Average B-factor | |
| Protein | 62.5 Å2 |
| Solvent | 49.6 Å2 |
| R.m.s.d. from ideal geometry | |
| Bond lengths | 0.006 Å |
| Bond angles | 0.987° |
Modeling the cis chromophore postrefinement
We modeled the cis chromophore after refining the structure because there was poor density for this conformation. There was, however, residual density in the region we expected the cis chromophore to be. We introduced the alternate conformation in COOT, fit it to the residual density, and ran the model through several rounds of PHENIX refinement, which resulted in the modeled positions shown in Figures 1 and 2 in turquoise.
Acknowledgments
The authors are grateful for the use of beamline 12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) in Menlo Park, CA, operated by Stanford University and supported by the Department of Energy and the National Institutes of Health. They additionally are thankful to Jens Kaiser and Pavle Nikolovski at the California Institute of Technology for helpful discussions. Finally, they thank the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program for support of the Molecular Observatory at the California Institute of Technology.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information Figure 1.
Supplementary Information Figure 2.
Supplementary Information Figure 3.
Supplementary Information Figure 4.
Supplementary Information Figure 5.
Supplementary Information Figure 6.
Supplementary Information Figure 7.
Supplementary Information Figure 8.
Supplementary Information Table and Figures Caption
References
- 1.Tromberg B, Shah N, Lanning R, Cerussi A, Espinoza J, Pham T, Svaasand L, Butler J. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia. 2000;2:26–40. doi: 10.1038/sj.neo.7900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikmi A, Gibson M. Identification and in vivo characterization of NvFP-7R, a developmentally regulated red fluorescent protein of Nematostella vectensis. PLoS One. 2010;5:e11807. doi: 10.1371/journal.pone.0011807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Jackson W, Steinbach P, Tsien R. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci USA. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shcherbo D, Merzlyak E, Chepurnykh T, Fradkov A, Ermakova G, Solovieva E, Lukyanov K, Bogdanova E, Zaraisky A, Lukyanov S, Chudakov D. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 5.Kredel S, Nienhaus K, Oswald F, Wolff M, Ivanchenko S, Cymer F, Jeromin A, Michel F, Spindler K-D, Heilker R, Nienhaus G, Wiedenmann J. Optimized and far-red-emitting variants of fluorescent protein eqFP611. Chem Biol. 2008;15:224–233. doi: 10.1016/j.chembiol.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Chica R, Moore M, Allen B, Mayo S. Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries. Proc Natl Acad Sci USA. 2010;107:20257–20262. doi: 10.1073/pnas.1013910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shcherbo D, Shemiakina I, Ryabova A, Luker K, Schmidt B, Souslova E, Gorodnicheva T, Strukova L, Shidlovskiy K, Britanova O, Zaraisky A, Lukyanov K, Loschenov V, Luker G, Chudakov D. Near-infrared fluorescent proteins. Nat Methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piatkevich K, Malashkevich V, Morozova K, Nemkovich N, Almo S, Verkhusha V. Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant. Sci Rep. 2013;3:1847. doi: 10.1038/srep01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shkrob M, Yanushevich Y, Chudakov D, Gurskaya N, Labas Y, Poponov S, Mudrik N, Lukyanov S, Lukyanov K. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochem J. 2005;392:649–654. doi: 10.1042/BJ20051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin M, McKeown M, Ng H-L, Aguilera T, Shaner N, Campbell R, Adams S, Gross L, Ma W, Alber T, Tsien R. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morozova K, Piatkevich K, Gould T, Zhang J, Bewersdorf J, Verkhusha V. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J. 2010;99:5. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jun C, Russell DH, Stéphane YC, Pengpeng L, Emilio G-G, John SB, Niloufar JA, Amy JL, Paula JC, Michelle AB, Michael WD, Ho-Leung N, Garcia KC, Christopher HC, Kang S, Helen MB, Michael ZL. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merzlyak E, Goedhart J, Shcherbo D, Bulina M, Shcheglov A, Fradkov A, Gaintzeva A, Lukyanov K, Lukyanov S, Gadella T, Chudakov D. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 14.Chan MCY, Karasawa S, Mizuno H, Bosanac I, Ho D, Prive GG, Miyawaki A, Ikura M. Structural characterization of a blue chromoprotein and its yellow mutant from the sea anemone Cnidopus japonicus. J Biol Chem. 2006;281:37813–37819. doi: 10.1074/jbc.M606921200. [DOI] [PubMed] [Google Scholar]
- 15.Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, Miller DJ, Wiedenmann J, Salih A, Matz MV. Diversity and evolution of coral fluorescent proteins. PLoS One. 2007;3:e2680. doi: 10.1371/journal.pone.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore M, Oteng-Pabi S, Pandelieva A, Mayo S, Chica R. Recovery of red fluorescent protein chromophore maturation deficiency through rational design. PLoS One. 2012;7:e52463. doi: 10.1371/journal.pone.0052463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strack R, Strongin D, Mets L, Glick B, Keenan R. Chromophore formation in DsRed occurs by a branched pathway. J Am Chem Soc. 2010;132:8496–8505. doi: 10.1021/ja1030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudalige K, Habuchi S, Goodwin P, Pai R, De Schryver F, Cotlet M. Photophysics of the red chromophore of HcRed: evidence for cis-trans isomerization and protonation-state changes. J Phys Chem B. 2010;114:4678–4685. doi: 10.1021/jp9102146. [DOI] [PubMed] [Google Scholar]
- 21.Pletnev S, Shcherbo D, Chudakov D, Pletneva N, Merzlyak E, Wlodawer A, Dauter Z, Pletnev V. A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis-trans isomerization of the chromophore. J Biol Chem. 2008;283:28980–28987. doi: 10.1074/jbc.M800599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam V, Lelimousin M, Boehme S, Desfonds G, Nienhaus K, Field M, Wiedenmann J, McSweeney S, Nienhaus G, Bourgeois D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc Natl Acad Sci USA. 2008;105:18343–18348. doi: 10.1073/pnas.0805949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loos D, Habuchi S, Flors C, Hotta J-I, Wiedenmann J, Nienhaus G, Hofkens J. Photoconversion in the red fluorescent protein from the sea anemone Entacmaea quadricolor: is cis-trans isomerization involved? J Am Chem Soc. 2006;128:6270–6271. doi: 10.1021/ja0545113. [DOI] [PubMed] [Google Scholar]
- 24.Wall M, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nature Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 25.Abbyad P, Childs W, Shi X, Boxer S. Dynamic Stokes shift in green fluorescent protein variants. Proc Natl Acad Sci USA. 2007;104:20189–20194. doi: 10.1073/pnas.0706185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu X, Wang L, Colip L, Kallio K, Remington SJ. Unique interactions between the chromophore and glutamate 16 lead to far-red emission in a red fluorescent protein. Protein Sci. 2009;18:460–466. doi: 10.1002/pro.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry. 2009;48:8279–8281. doi: 10.1021/bi900870u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suto K, Masuda H, Takenaka Y, Tsuji FI, Mizuno H. Structural basis for red-shifted emission of a GFP-like protein from the marine copepod Chiridius poppei. Genes Cells. 2009;14:727–737. doi: 10.1111/j.1365-2443.2009.01305.x. [DOI] [PubMed] [Google Scholar]
- 29.Kredel S, Oswald F, Nienhaus K, Deuschle K, Röcker C, Wolff M, Heilker R, Nienhaus GU, Wiedenmann J. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One. 2008;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nienhaus K, Nar H, Heilker R, Wiedenmann J, Nienhaus GU. Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611. J Am Chem Soc. 2008;130:12578–12579. doi: 10.1021/ja8046443. [DOI] [PubMed] [Google Scholar]
- 31.Adams P, Afonine P, Bunkóczi G, Chen V, Davis I, Echols N, Headd J, Hung L-W, Kapral G, Grosse-Kunstleve R, McCoy A, Moriarty N, Oeffner R, Read R, Richardson D, Richardson J, Terwilliger T, Zwart P. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of Coot. Acta Cryst D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Figure 1.
Supplementary Information Figure 2.
Supplementary Information Figure 3.
Supplementary Information Figure 4.
Supplementary Information Figure 5.
Supplementary Information Figure 6.
Supplementary Information Figure 7.
Supplementary Information Figure 8.
Supplementary Information Table and Figures Caption


