Figure 4.
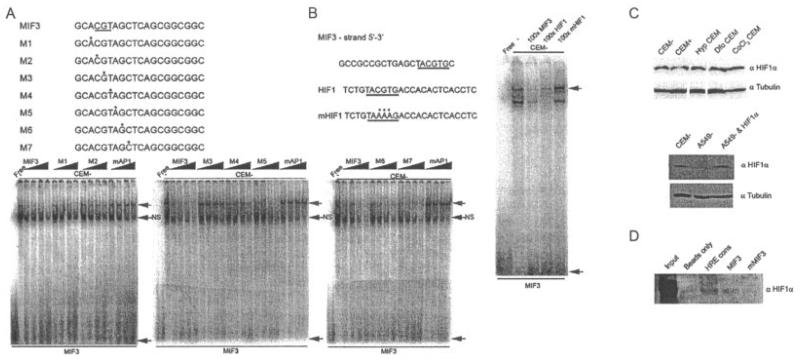
The MIF-3 complex is composed of hypoxia-inducible factor 1α (HIF-1α). A, Electrophoretic mobility shift assays (EMSAs) of nuclear extracts prepared from untreated CEMC7A cells (CEM−) incubated with radiolabeled oligonucleotides corresponding to MIF-3. Competition experiments using increasing concentrations (5×−100×) of a series of probes (M1–M7) containing purine-pyrimidine substitutions were performed. Point mutations of bases in the oligonucleotides M2, M3, and M4 resulted in loss of competition. B, EMSA of nuclear extracts prepared from untreated CEMC7A cells incubated with radiolabeled oligonucleotide corresponding to MIF-3. Competition experiments were performed by adding unlabeled oligonucleotides as indicated. In A and B, asterisks show mutated bases in oligonucleotides used in the competition experiments. Nucleotides required for binding are underlined. Shaded arrows show sequence-specific complexes; solid arrows show free probe. NS = nonspecific complex. C, Immunoblots of nuclear extracts prepared from untreated CEMC7A cells, CEMC7A cells treated with 1 μM DEX for 20 hours (CEM+), CEMC7A cells grown under hypoxic (Hyp) conditions for 20 hours, CEMC7A cells treated with 100 μM desferrioxamine (Dfo) for 20 hours, CEMC7A cells treated with 100 μM CoCl2 for 20 hours, untreated A549 cells (A549–), and A549 cells transfected with HIF-1α expression vector. D, DNA affinity chromatography using CEMC7A nuclear extract and the HIF response element consensus (HRE cons), MIF-3, or mutated MIF-3 (mMIF-3) sequences. Bound protein–DNA complexes were subjected to immunoblotting. Results are representative of at least 3 separate experiments. See Figure 1 for other definitions.