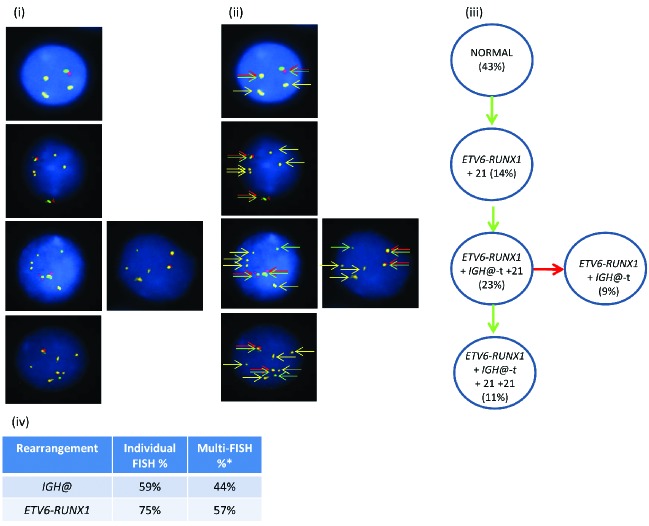
Figure 2.
Images of FISH interphase nuclei depicting the apparent emergence of an ETV6-RUNX1 translocation prior to the IGH@-t. (i) Captured interphase nuclei from patient 20951 hybridized with FISH probes to track the IGH@-t (red/green) and the ETV6-RUNX1 translocation (homemade RUNX1 probe; gold). (ii) The same captured interphase nuclei, including colored arrows highlighting the signals that were scored: the yellow arrows highlight the gold probe marking the RUNX1 rearrangement and the red/green double arrows highlight the IGH@ break apart probe that marks both the copy number of chromosome 14 and also detects the presence of a translocation. (iii) Cartoon depicting possible models of evolutionary progression either by gain (green arrows) or loss (red arrows). Forty-three percent of nuclei were normal for both probes (one red/green fusion appears yellow, this is marked with arrows for clarity as shown in (ii) with 14% showing an additional copy of chromosome 21 in the presence of the RUNX1 translocation. On the background of the RUNX1 translocation and additional chromosome 21, a sub-clone acquired an IGH@-t (23%) which subsequently either lost (9%) or gained (11%) a copy of chromosome 21. (iv) Table showing the percentage of positive nuclei when samples were hybridized with either the individual probes for each rearrangement (Individual FISH), or when they were investigated together (Multi-FISH) in the same cell.