Figure 4.
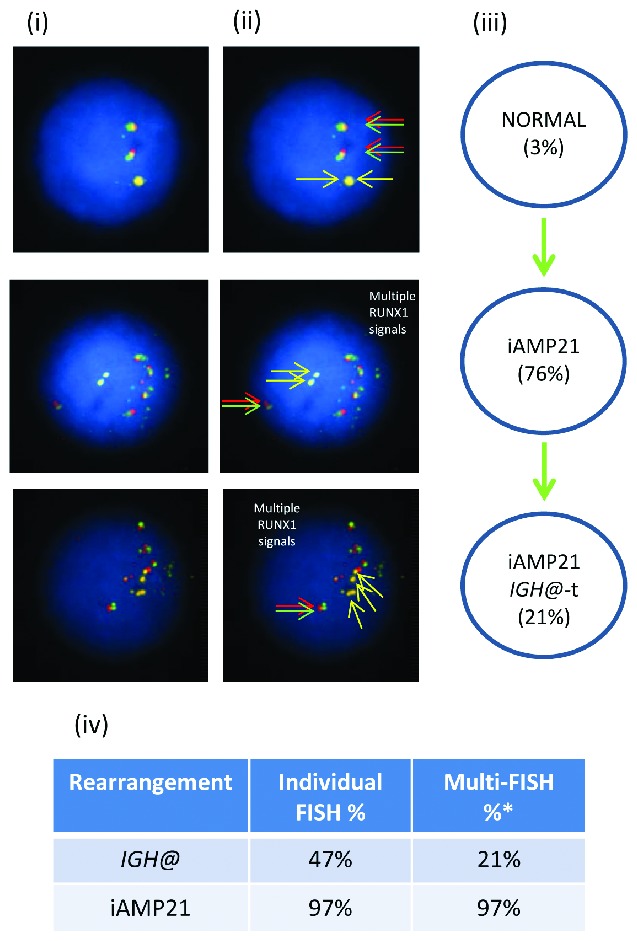
Images of FISH interphase nuclei depicting the apparent emergence of an iAMP21 prior to the IGH@-t. (i) Captured interphase nuclei from patient 11061 hybridized with FISH probes to track iAMP21 using a home-made RUNX1 breakapart probe (red/green) and a home-made IGH@ single color probe (gold). (ii) The same captured interphase nuclei, including colored arrows highlighting the signals that were scored: the yellow arrows highlight the gold probe marking the home-made IGH@ probe that detects both the copy number of chromosome 14 and also the presence of a translocation. The red/green double arrows highlight the home home-made RUNX1 breakapart probe that marks the iAMP21 aberration. (iii) Cartoon depicting a possible model of evolutionary progression. Seventy-six percentage of nuclei showed multiple copies of RUNX1 with 21% of these nuclei also harboring IGH@-t. (iv) Table showing the percentages of positive nuclei when samples were hybridised with either the individual probes for each rearrangement (Individual FISH), or when they were investigated together (Multi-FISH) in the same cell.
