Abstract
Id proteins (Id1-Id4) are helix-loop-helix (HLH) transcriptional regulators that lack a basic DNA binding domain. They act as negative regulators of basic helixloop-helix (bHLH) transcription factors by forming heterodimers and inhibit their DNA binding and transcriptional activity. Id proteins are implicated in the regulation of various cellular mechanisms such as cell proliferation, cellular differentiation, cell fate determination, angiogenesis and tumorigenesis. A handful of recent studies also disclosed that Id proteins have critical functions in adipocyte differentiation and adipose tissue metabolism. Here, we reviewed the progress made thus far in understanding the specific functions of Id proteins in adipose tissue differentiation and metabolism. In addition to reviewing the known mechanisms of action, we also discuss possible additional mechanisms in which Id proteins might participate in regulating adipogenic and metabolic pathways.
Keywords: WAT, BAT, Thermogenesis, Insulin Sensitivity, Lipid Metabolism, Review
2. INTRODUCTION
Obesity is rapidly emerging as one of the most serious health problems worldwide since its prevalence has drastically increased over the past 3 decades, reaching epidemic proportions (1, 2). A number of factors such as energy rich diets, sugary drinks, lack of physical exercise and technology-driven sedentary life styles have contributed to the current levels of obesity. Obesity is associated with an array of diseases and is an established risk factor for a number of diseases such as hypertension, type 2 diabetes, dyslipidemia, chronic heart disease, nonalcoholic fatty liver disease (NAFLD), atherosclerosis, degenerative disorders, including dementia and airway diseases (3–6). Moreover, various epidemiological and clinical studies suggest that obesity is an independent risk factor for various cancers such as liver, colon, breast, gastric, gall bladder, endometrial, esophagus and pancreatic cancers and renal cell carcinoma (7–10). Therefore, understanding the development, maintenance and function of fat (adipose) tissue, and in particular, the molecular mechanisms that govern adipocyte differentiation and expansion of adipose tissues and their metabolic functions in normal and obese conditions have become increasingly essential for the treatment and intervention of obesity.
Two types of functionally distinct adipose tissue exist in the body: white adipose tissue (WAT) and brown adipose tissue (BAT) (11). The major function of WAT is to store excess energy in the form of triglycerides when energy intake exceeds expenditure. The adipocyte number, size, and the total amount of WAT are dynamically in proportion to the energy levels of the body (12). If the energy influx is consistently higher than expenditure, it leads to continuous synthesis of triglycerides, with a concomitant increase in adipocyte size (hypertrophy) due to triglyceride accumulation. In addition, these excessive triglyceride levels in the body force preadipocytes and/or adipocyte progenitors in the WAT to differentiate into adipocytes and store newly synthesized triglycerides. This leads to a steady increase in total body WAT, resulting in overweight and ultimately obesity. In recent years, significant progress has been made in understanding the development, differentiation, maintenance and specific functions of adipose tissues. A number of genes, such as members of the PPAR and C/EBP, and KLF, STAT, SREBP-1c, FOXC2, E2F, Rb, Wnt, and GATA have been identified to play essential roles in regulating white adipocyte differentiation and metabolic function (13–17). Deletion of some of these genes in mice resulted in a lean phenotype due to impaired adipogenesis, altered adipose tissue metabolism and reduction in total body WAT.
In contrast to WAT, BAT is specialized for energy expenditure by dissipating energy as heat, a process termed adaptive thermogenesis. This unique metabolic property of production of heat by BAT is attributed to its high mitochondrial density and its exclusive expression of uncoupling protein-1 (UCP1) in the inner mitochondrial membrane. During respiration, an electron-motive force established across the inner mitochondrial membrane is dissipated as heat by UCP1, rather than being used to drive the synthesis of ATP. For the past few years BAT has been receiving tremendous interest due to its ability to expend energy and function as a defense against hypothermia and possibly obesity. Subsequently, a number of genes such as PGC1α, UCP1, PPARγ, BMP, EBF2, Cidea, FOXC2, SRC2, PRDM16, Orexin, LXR, ARRDC3, Twist1, and TRα1 were discovered that play crucial roles in BAT development, maintenance and its thermogenesis function (18–24). PGC1α especially has emerged as the master regulator of various genes that are involved in thermogenesis, and it predominantly controls the entire BAT-mediated thermogenesis program (24). On the other hand, BMP and Ebf2 determine between the white versus brown adipose progenitor differentiation programs (25, 26). In addition to the above genes, recent studies revealed the involvement of another family of proteins, inhibitor of DNA binding (Id), in adipocyte differentiation and adipose tissue metabolism.
3. Id PROTEINS
Id proteins (Id1, Id2, Id3 and Id4) are a subfamily of helix-loop-helix (HLH) transcription factors that lack a basic DNA binding domain. Therefore, they function entirely by dimerization with other transcriptional regulators, mainly those of the basic-helix-loop-helix (bHLH) factors. The heterodimers (Id/bHLH) fail to bind to DNA, and hence Id proteins function as dominant negative regulators of bHLH proteins (27). The Id proteins (Id1-Id4) range in size from 120–160 amino acids and, despite the highly conserved HLH domain among all 4 Id proteins, they display extensive sequence divergence. Expression analysis of Id proteins revealed widespread and overlapping expression patterns in multiple tissues, suggesting that they play an essential role in many cell types with the existence of possible redundancy in their functions. A number of in vitro and in vivo studies implicated Id proteins in the regulation of multiple cellular processes such as cell cycle regulation, cell proliferation, cellular differentiation, cell fate determination, hematopoiesis, angiogenesis and tumorigenesis. As a general mechanism of action, Id proteins induce their inhibitory effect by acting as negative regulators of basic helix-loop-helix (bHLH) transcription factors, which control cell type-specific gene expression. They form heterodimers with DNA binding bHLH proteins and prevent their DNA binding and transcriptional activity. For example, Id1 regulates the transcription of the cell cycle inhibitor p16 by directly binding to its transcriptional activators E47 and Ets2 (Figure 1A). Some of the well-known targets of Id proteins include E proteins, Rb, p107, p130, PAX, Ets, MyoD and Myf-5 (28–31). The specific function of Id proteins and their mechanism of action in cell cycle control, cellular differentiation, hematopoiesis and tumorigenesis have been extensively reviewed elsewhere (28–34). Here, we review the progress made thus far in understanding the specific functions of the Id family of proteins in adipogenesis and adipose tissue metabolism. Since the mechanism of action of Id proteins in cellular metabolism is still an active area of investigation, in addition to reviewing their known mechanisms of action in adipose tissue metabolism, we also provide our views on possible additional mechanisms in which Id proteins might participate to regulate adipogenic and metabolic pathways.
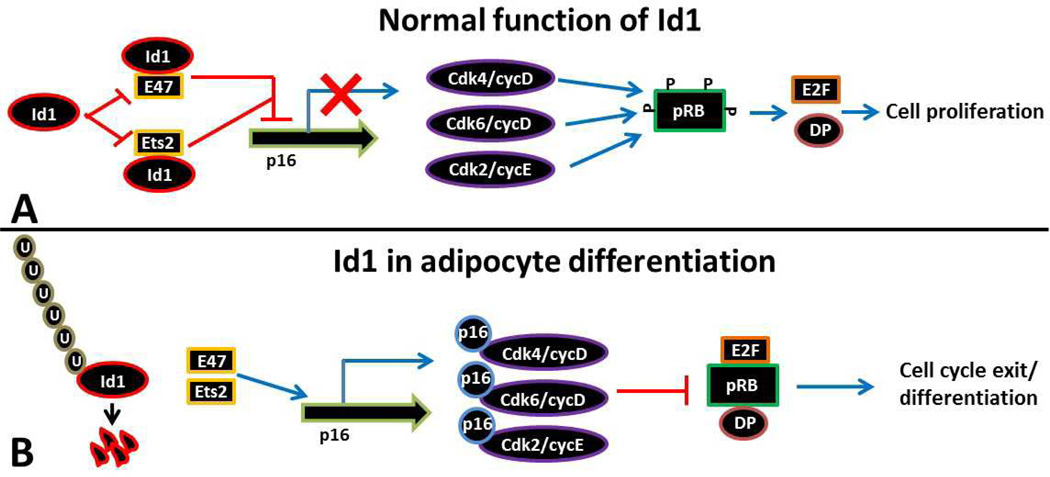
Figure 1.
(A). Transcriptional regulation of p16 by Id1 through E47 and Ets2, leading to cell proliferation. Id1 prevents the expression of the cell cycle inhibitor p16 through E47 and Ets2. In the absence of p16 the uninhibited Cdk/cyclin complexes continue to promote cell cycle progression by phosphorylating and keeping pRB in its inactive state. This leads to constitutive activation of E2F and DP factors, which activate a number of S phase genes that drive cell proliferation (29, 30, 33, 34). (B). During adipocyte differentiation, Id1 is ubiquitinated and targeted for degradation. We speculate that this facilitates expression of p16 through E47 and Ets2. Subsequently, p16 inhibits Cdk/cyclin complexes which fail to phosphorylate pRB leading to cell cycle exit and adipocyte differentiation.
3.1. Id1
A protein expression screen in different metabolic tissues of adult mice revealed higher expression of Id1 in both WAT and BAT. Id1 expression is especially highest in BAT compared to other metabolic organs (35). These observations suggest that Id1 could possibly play a role in adipogenesis, WAT metabolism and BAT-mediated thermogenesis. Subsequently, when wild-type mouse embryonic fibroblasts (MEFs) or 3T3-L1 preadipocytes were induced to differentiate into adipocytes, Id1 protein was strongly expressed before the induction of differentiation, but rapidly declined and completely disappeared during adipocyte differentiation (35). It appears that the clearing of Id1 protein from cells might be necessary before the cells start differentiating into adipocytes. Why is it necessary to clear Id1 from the cells to initiate differentiation? One possibility could be that Id1 promotes cell proliferation and inhibits cellular differentiation (28, 29). Id1 prevents the expression of the cell cycle inhibitor p16 through E47 and Ets2 (36, 37). Id1 can sequester the bHLH transcription factor E47, which is a transcriptional activator of p16. In addition, phosphorylation of the Ets family transcription factor Ets2 by Ras/Raf/MEK signaling leads to transcriptional activation of p16. However, expression of p16 by Ets2 can be effectively inhibited by Id1 through its direct interaction with Ets2 (27, 37, 38). In the absence of p16 the uninhibited Cdk/cyclin complexes continue to promote cell cycle progression by phosphorylating and keeping pRB in its inactive state, leading to constitutive activation of E2F and DP factors, which activate a number of genes that drive cell proliferation (Figure 1A). It is undesirable to initiate cellular differentiation because in order for the cells to differentiate into adipocytes, they need to permanently exit from the cell cycle and reach a state of irreversible growth arrest for terminal differentiation. In the presence of Id1, cells may not reach this growth arrest stage due to its ability to block p16 expression, leading to constitutive activation of Cdk/cyclin complexes followed by pRB inactivation, which continues to promote cell proliferation. This is further supported by the observation that pRB−/− cells fail to differentiate into adipocytes, and repression of E2F transcription by pRB contributes to establishing a permanent exit from the cell cycle (39). Furthermore, it was demonstrated that C/EBPα promotes adipocyte differentiation by repressing E2F–dependent transcription. However, C/EBPα mutants defective in repression of E2F–dependent transcription have an impaired ability to suppress proliferation and thereby fail to induce adipocyte differentiation (40). This further indicates that inhibition of E2F–mediated cell proliferation is a pre-requisite for the initiation of adipocyte differentiation. This could be the primary reason why Id1 is targeted for degradation and cleared from the cells, which facilitates p16 expression, and ultimately, E2F–repression, which is required to initiate adipocyte differentiation (Figure 1B). In line with these studies, in the absence of Id1, the ability of Id1−/− MEFs to differentiate into adipocytes was significantly accelerated. It appears that p16-mediated inhibition of Cdk/cyclin complexes in the absence of Id1 helped cells to exit the cell cycle, which is essential for differentiation. As a result, earlier expression of PPARγ, the master regulator of adipocyte differentiation, and its downstream target, aP2 were detected in differentiating Id1−/− cells (35).
Alternatively, it is also possible that Id1 regulates PPARγ and operates upstream to PPARγ. Since PPARγ expression is induced in the absence of Id1, overexpression of Id1 could directly demonstrate whether PPARγ is really downstream to Id1. If that is the case, overexpression of Id1 should suppress the expression and/or activity of PPARγ and its downstream target genes such as aP2, CD36, perilipin, lipoprotein lipase (LPL) and phosphoenol pyruvate carboxykinase (15, 41), which in turn should delay or block adipogenesis. However, surprisingly, Id1-overexpressing 3T3-L1 preadipocytes showed the same degree of adipocyte differentiation as control cells. This is because, in addition to endogenous Id1, overexpressed Id1 was also rapidly cleared from cells during adipocyte differentiation. This further demonstrates how essential it is to clear all the Id1 from cells to facilitate initiation of adipocyte differentiation. Id1 was heavily ubiquitinated and targeted for proteasome-mediated degradation in Id1-overexpressing 3T3-L1 cells undergoing differentiation. As a result, the overexpressed Id1 was unable to inhibit adipocyte differentiation as expected (35). Therefore, it is unclear whether Id1 really operates upstream to PPARγ, and if so, how it regulates PPARγ. It is possible to modify ubiquitin-binding lysine sites in Id1 protein and reevaluate if the mutant Id1 suppresses adipogenesis. However, it was shown before that generation of a lysine-less Id1, where all the lysine residues were replaced to alanine, still could not prevent Id1 from degradation, and Id1 underwent N-terminus-dependent ubiquitination which is modulated by MyoD (42). During adipocyte differentiation, in addition to N-terminus-dependent ubiquitination and degradation, Id1 could also have undergone ubiquitin-mediated degradation by other E3 ligases such as Smurf2. A recent study showed that Smurf2 mediates Id1 degradation in senescent cells (43). Another potential problem with the approach of lysine residue replacement is that amino acid substitutions in Id1 protein could abolish Id1’s transcriptional regulatory functions, and this mutant Id1 may no longer sequester its target bHLH factors as effectively as the native Id1. Nevertheless, the studies in Id1−/− cells at least established the basic function of Id1 in adipogenesis, and in the absence of Id1, adipocyte differentiation is accelerated with earlier expression of PPARγ and its downstream target genes.
Surprisingly, although adipogenesis is accelerated, Id1 knockout mice are leaner compared to wild-type controls. This lean phenotype is especially more striking in aged animals as the Id1−/− mice failed to gain fat mass during aging (35). Since adipogenesis is not defective in the absence of Id1, a failure to gain fat mass during aging in Id1−/− mice could be due to increased energy expenditure. This speculation is mainly fueled by the fact that Id1 expression is strongest in BAT compared to other metabolic tissues, and hence, Id1 might have a significant role in the regulation of BAT-mediated thermogenesis. Consistent with this prediction, the expression levels of PGC1α and its downstream target UCP1 are up-regulated in Id1−/− BAT at room temperature and in response to cold-induced thermogenesis, suggesting that Id1 deficiency resulted in increased thermogenesis. This is further strengthened by the observation that Id1−/− mice exhibited higher O2 consumption and lower respiratory exchange rate (RER), indicating that energy expenditure is increased in Id1-deficient mice and that they use a relatively higher proportion of lipid as an energy source compared to control mice (35). Thus, the failure to gain fat mass with age in Id1−/− mice could be explained, in part, by an increase in lipid oxidation and energy expenditure. PGC1α is the master regulator of thermogenesis and controls the entire BAT-mediated thermogenesis program (24). Although PGC1α levels are increased in the BAT in the absence of Id1, it is unclear how Id1 regulates PGC1α expression and/or its transcriptional activity. Since Id1 acts as a dominant negative regulator of other transcription factors, one possibility could be that Id1 may directly bind to PGC1α and suppress its transcriptional activity. Alternatively, Id1 may co-operate with other negative regulators of PGC1α such as Rb and Twist1, thereby controling the expression and activity of PGC1α. In the absence of Id1 where there is less inhibitory environment for PGC1α, it is relatively free to induce transcriptional activation of its downstream targets such as UCP1 at a higher rate, thereby increasing thermogenesis. However, additional in vitro and in vivo studies are required to more clearly understand the transcriptional inhibitory functions of Id1 in the thermogenesis signaling pathway. Especially since Id1 is one of the major regulators of E, Ets, Rb, and PAX proteins, these studies should be directed at understanding how Id1/E, Id1/Ets or Id1/pRB protein signaling regulate the expression and activities of the PGC1α network of proteins involved in thermogenesis.
3.2. Id2
Induction of adipogenesis in the 3T3-F442A preadipocyte cell line by the adipogenic compound harmine followed by transcriptional profiling identified Id2 as a potential regulator of adipocyte differentiation since its expression is induced during differentiation (44). Accordingly, overexpression of Id2 increased the ability of preadipocytes to differentiate into adipocytes with induced expression of PPARγ and its downstream target genes aP2, CD36, LPL, adiponectin, and C/EBPα. Conversely, knockdown of Id2 by siRNA inhibited the ability of preadipocytes to differentiate into adipocytes due to impaired induction of PPARγ and its downstream target genes (44). This suggests that Id2 might be directly regulating PPARγ. Accordingly, overexpressed Id2 increased the expression of PPARγ, whereas either overexpression or knockdown of PPARγ did not influence the expression of Id2. These studies indicate that PPARγ does not regulate Id2, but PPARγ expression is regulated by Id2, and PPARγ operates downstream to Id2. In contrast, inhibition of Id2 had no effect on the induction of other transcription factors such as C/EBPβ and C/EBPδ, which also function to activate PPARγ in a parallel adipocyte differentiation pathway. This is perhaps not surprising since Id2 is a direct downstream target of C/EBPβ, and C/EBPβ can directly bind to the Id2 promoter and induce its expression (45). This explains why inhibition of adipocyte differentiation is only partial when Id2 is knocked down since C/EBPβ and C/EBPδ are still able to activate PPARγ and drive adipocyte differentiation in the absence of Id2.
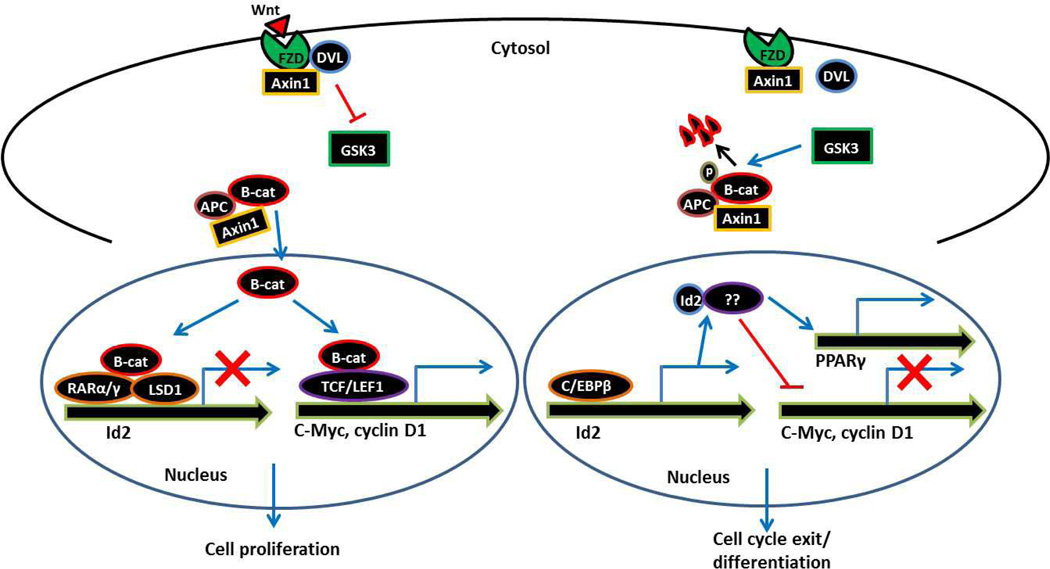
In this context, it is essential to understand which receptor/ligand signaling pathway regulates Id2, and subsequently, how Id2 controls PPARγ during adipocyte differentiation. It appears that Id2 expression could be regulated by Wnt-β-catenin signaling. Treatment of preadipocytes with Wnt-3a–conditioned media not only suppressed PPARγ expression but also Id2 expression, suggesting that Wnt signaling regulates Id2 expression during adipocyte differentiation (44). Wnt signaling is initiated when Wnt ligands bind to membrane receptors of the Frizzled family, leading to Dishevelled-mediated inhibition of kinase activity of a complex containing glycogen synthase kinase 3 (GSK3), β-catenin, Axin1 and APC (46, 47). Due to GSK3 inhibition in the presence of Wnt, β-catenin cannot be phosphorylated and targeted for degradation, and the hypophosphorylated, stabilized β-catenin translocates to the nucleus and binds to the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors. TCF/LEF-β-catenin complexes bind to TCF/LEF-binding sites or Wnt-response elements (WRE) in the promoters of target genes such as c-Myc and cyclin D1 leading to their transcriptional activation. Therefore, active Wnt signaling promotes the maintenance of a proliferative state and inhibits differentiation (Figure 2, Left) (46, 47). For example, Wnt-1-expressing cells fail to express C/EBPα and PPARγ, which are required for adipocyte differentiation (16). Does Wnt-β-catenin signaling directly regulate Id2 expression? Although such a mechanism was not investigated thus far in the differentiated adipocytes, it was demonstrated in human keratinocytes that β-catenin, indeed, directly suppresses Id2 expression. β-catenin directly binds to WRE at the Id2 promoter and recruits retinoic acid receptors (RARs) to the WRE at the Id2 promoter. This is followed by RAR-dependent recruitment of LSD1 demethylase to the WRE in the Id2 promoter, leading to a reduction in histone H3 and H4 acetylation and histone H3 K-4 methylation, resulting in transcriptional repression (48). Similar mechanism might operate in adipocytes and therefore blocking Wnt signaling is not only necessary for activation of Id2 but also for the C/EBPs, which collectively induce PPARγ and initiate adipocyte differentiation (Figure 2, Right). Consistent with this hypothesis, inhibition of Wnt signaling in 3T3-L1 cells by dominant-negative TCF4 (dnTCF4) induced adipogenesis, suggesting that blocking Wnt signaling is necessary to initiate adipocyte differentiation (16). Thus, the Wnt-β-catenin pathway appears to regulate both the C/EBP and Id2 parallel pathways that lead to activation of PPARγ and adipocyte differentiation. However, although how the C/EBPs regulate PPARγ is well established (49), the precise mechanism by which Id2 regulates PPARγ is largely unclear, and further studies are required to establish a molecular link between Id2 and PPARγ.
Figure 2.
Possible mechanism showing the regulation of Id2 in the presence or absence of active Wnt signaling and consequently how Id2 regulates adipocyte differentiation through PPARγ. (Left) In the presence of Wnt, β-catenin cannot be phosphorylated and targeted for degradation. β-catenin translocates to the nucleus and performs two tasks: 1. Binds to the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors. TCF/LEF-β-catenin complexes bind to Wnt-response elements (WRE) in the promoters of target genes such as c-Myc and cyclin D1 leading to their transcriptional activation. Thus, active Wnt signaling promotes cell proliferation and inhibits adipocyte differentiation (46, 47). 2. β-catenin could suppress Id2 expression by directly binding to WRE at the Id2 promoter and recruits retinoic acid receptors (RARs) to the WRE at the Id2 promoter. This is followed by RAR-dependent recruitment of LSD1 demethylase, leading to a reduction in histone H3 and H4 acetylation and histone H3 K-4 methylation, resulting in Id2 transcriptional repression and impaired adipogenesis (48). (Right) The precise mechanism by which Id2 regulates PPARγ is not known. One possibility could be that in the absence of Wnt, C/EBPs might gain access to the Id2 promoter and induce its expression. Subsequently, Id2 by directly binding to an unknown factor, on one hand induces PPARγ expression and on the other hand suppresses c-Myc and cyclin D1 expression leading to cell cycle exit and adipocyte differentiation.
In accordance with these observations in the preadipocyte cell lines, 4–6-day-old Id2−/− neonates exhibited detectable deficiencies in interscapular and inguinal WAT tissues. Similarly, Id2−/− MEFs subjected to adipocyte differentiation exhibited a partial impairment in adipogenic potential with concomitant reduction in the levels of PPARγ and its downstream targets, LPL, adiponectin, and aP2 (44). A partial reduction in the adipocyte differentiation potential of Id2−/− cells indicate that although C/EBPβ and C/EBPδ can still activate PPARγ and drive adipocyte differentiation, Id2 operates in a parallel pathway and is required for complete activation of PPARγ, leading to normal differentiation. In vivo, Id2 expression was significantly elevated in the adipose tissues of mice fed a high-fat-diet (HFD) as well as genetically obese mice. However, whether impaired adipogenesis in the absence of Id2 can lead to an anti-obesity phenotype similar to that of C/EBPα and PPARγ-deficient mice is unknown. This is mainly because, although the gross phenotype of Id2−/− mice at birth was indistinguishable from wild-type litter mates, after day 6, Id2−/− neonates show severe growth defects and ~80% of the mice do not survive beyond 2–3 weeks of age. Only ~20% of mice escape from death but they are severely growth retarded (44, 50). These Id2−/− mice displayed several metabolic phenotypes such as increased glucose tolerance, insulin sensitivity, reduced gonadal WAT, altered daily and circadian rhythms of feeding and locomotor activity, and increased energy expenditure (51). Although some of the observed metabolic phenotypes could be attributed to severe growth defects in Id2−/− mice, it cannot be excluded that lack of Id2 could also have direct impact on adipose tissue metabolism. This is due to the fact that Id2 can directly regulate adipocyte determination and differentiation factor 1 (ADD1), also known as sterol regulatory element-binding protein 1c (SREBP-1c). SREBP-1c, a basic helix-loop-helix leucine-zipper (bHLH-LZ) transcription factor, regulates the expression of various genes involved in fatty acid and triglyceride metabolism and is a potent trans-activator of the fatty acid synthase (FAS) promoter. Id2 can directly bind to SREBP-1c and inhibit it from transactivating FAS, thereby regulating fatty acid synthesis and metabolism (52). Moreover, SREBP-1c can also regulate a number of genes such as glycerol-3-phosphate acyltransferase, S14, stearoyl CoA desaturase, adiponectin and leptin. Therefore, in the absence of Id2, regulation of SREBP-1c as well as its target genes could be altered, leading to dramatic changes in adipose tissue metabolism. However, additional studies, such as specific deletion of Id2 in the adipose tissues by crossing Id2fl/fl mice with aP2Cre mice followed by metabolic analysis, are required in order to clearly understand the specific role of Id2 in adipose tissue maintenance and metabolism in vivo.
3.3. Id3
The role of Id3 in adipocyte differentiation and adipose tissue metabolism created considerable debate due to recent studies that conflicted with previous reports. One of the studies described that Id3 mRNA levels are abundant in proliferating 3T3-F422A preadipocytes but rapidly declined during adipocyte differentiation. Subsequently, forced expression of Id3 in preadipocytes prevented adipocytes from differentiation, as measured by adipsin and glycerol phosphate dehydrogenase activity, markers of adipocyte differentiation (53). However, a recent study demonstrated that overexpression of Id3 did not result in any detectable changes in the expression of GLUT4, another marker of adipocyte differentiation. They also did not detect any significant differences in Oil-Red-O lipid staining between control and Id3-overexpressing cells or between wild-type and Id3−/− MEFs that were induced to differentiate into adipocytes, suggesting that either overexpression or lack of Id3 did not alter the adipocyte differentiation program (54). Timing of analysis and the choice of adipocyte markers might have partially contributed to these conflicting observations in these two studies. Further studies in Id3−/− mice also did not reveal any significant changes in the metabolic phenotypes when they were fed a regular diet compared to wild-type mice. Nevertheless, when the mice were fed a HFD, expansion of visceral adipose depots was significantly reduced in Id3−/− mice, whereas, such a reduction was not observed in other WAT locations (54). Accordingly, Id3 expression was more abundant in visceral compared to subcutaneous WAT, and Id3 expression was elevated in response to HFD only in visceral but not in subcutaneous adipose tissues of wild-type mice. Due to impaired visceral WAT expansion, Id3−/− mice were protected from HFD-induced obesity (54). However, no detectable differences were observed in the expression pattern of adipocyte differentiation markers such as aP2 or C/EBPα in visceral adipocytes, indicating that reasons other than defects in adipocyte differentiation are primarily responsible for the observed reduction in visceral adiposity in Id3−/− mice.
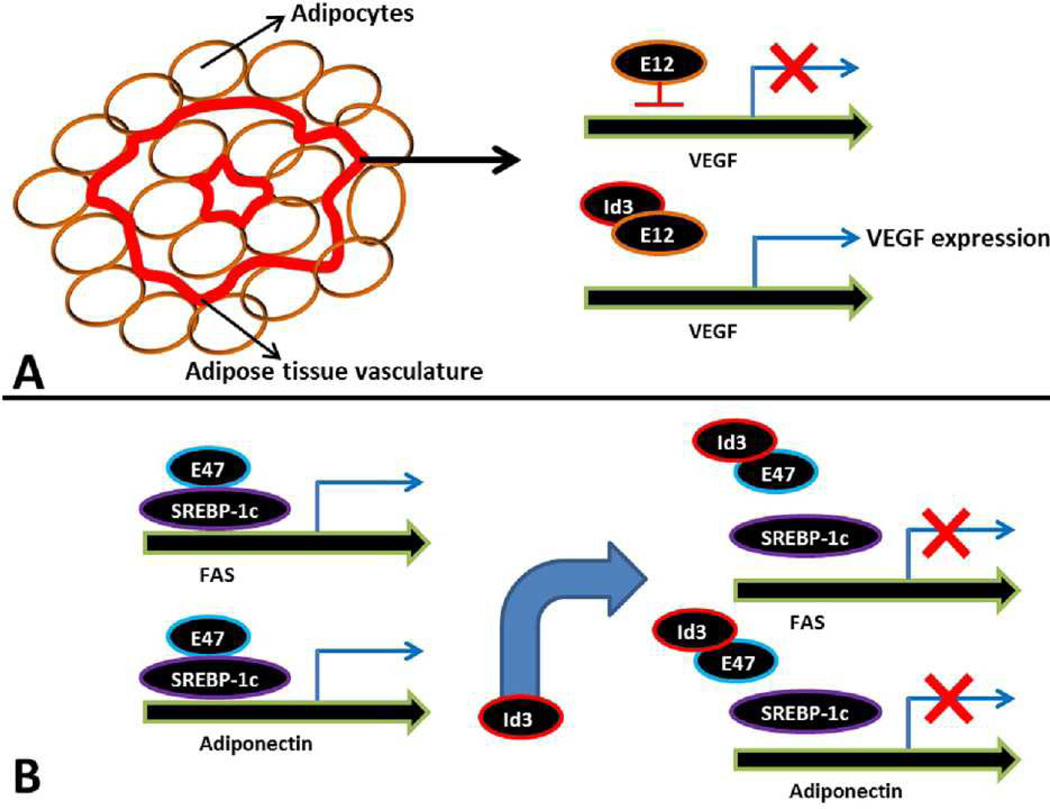
To explore the possible reason behind impaired visceral adipose tissue expansion in response to HFD, expression analysis of Id3 in adipocytes and stromal vascular fraction (SVF) of visceral adipose tissues in wild-type mice was performed. This analysis revealed induced expression of Id3 only in SVF but not in adipocytes in response to HFD, suggesting that a defect in visceral adipose tissue vascularization might have contributed to impaired visceral adipose tissue expansion in Id3−/− mice. Consistent with this hypothesis, expression of the angiogenesis factor VEGF is significantly induced in the visceral adipose tissue of wild-type mice in response to HFD but not in Id3−/− mice. This suggests that adipose tissue vascularization is impaired in the visceral adipose tissue of Id3−/− mice, leading to a failure in the expansion of adipose tissue (54). Previous studies have demonstrated that lack of Id3 leads to impaired angiogenesis (55), and therefore it is not entirely surprising that Id3 also plays a similar role in adipose tissue angiogenesis during its expansion. However, an important question that needs to be resolved is how does Id3 regulate VEGF expression? The VEGF promoter consists of several E-boxes where the bHLH factors bind and regulate its expression (56). The authors showed that transient expression of the E-protein E12 suppresses the expression of VEGF, indicating that E12 functions as a transcriptional repressor of VEGF. Id3 may promote VEGF expression by interacting with and preventing E12 from binding to the VEGF promoter (Figure 3A). Consistent with this idea, Id3 significantly antagonized the suppression of VEGF by E12 (54). This explains why there is a defect in the visceral adipose tissue vascularization and adipose tissue expansion in the absence of Id3. Taken together, these results suggest that loss of Id3 attenuates visceral fat expansion by inhibiting HFD-induced visceral fat VEGF expression and growth of blood vessels. However, E proteins often function as activators of gene expression, and it is somewhat intriguing that E12 functions as a transcriptional repressor of VEGF and Id3 binds to and releases this repression during adipose tissue vascular growth. It will be interesting to study if Id3 or its other family members are also involved in the regulation of other angiogenic factors during adipose tissue expansion.
Figure 3.
(A) Possible mechanism describing the role of Id3 in adipose tissue vascularization during adipose tissue expansion. VEGF is essential for growth of blood vessels during visceral fat expansion. E12 functions as a transcriptional repressor of VEGF. Id3 promotes VEGF expression by interacting with and preventing E12 from binding to the VEGF promoter (54). (B). Possible mechanism showing the role of Id3 in adipose tissue-associated fatty acid and adipokine metabolism. E47 interacts with SREBP-1c, a positive regulator of FAS and adiponectin, and enhances SREBP-1c–mediated promoter activation. Id3 regulates SREBP-1c activity indirectly by interacting with E47. Id3/E47 interaction prevents E47 binding to the SREBP-1c leading to impaired activation of FAS and adiponectin promoters and their expression (58).
In addition to Id3’s role in adipocyte differentiation, its role in adipose-specific metabolism is further complicated by conflicting reports on the regulation of SREBP-1c by Id3. SREBP-1c is a member of the bHLH family of transcription factors but also contains a leucine zipper (bHLH-LZ). SREBP-1c functions as a transcriptional activator of a number of genes involved in fatty acid and adipokine metabolism, such as FAS, leptin and adiponectin (57). Based on their structure, whether HLH-LZ factors can directly bind to Id proteins is debatable. A previous study showed that Id3 and SREBP-1c interact with each other, thereby Id3 antagonizes SREBP-1c transactivation of the FAS promoter. The authors were able to co-immunoprecipitate Id3 and SREBP-1c in a cell-free system. Moreover, in an in vitro translated system they demonstrated that Id3 alters SREBP-1c binding to the FAS promoter (52). However, later studies in a mammalian 2-hybrid system or by co-immunoprecipitation were unable to identify such a direct interaction between Id3 and SREBP-1c (58). Instead, they discovered that Id3 inhibits SREBP-1c through the E-protein E47. E47 interacts with SREBP-1c, a positive regulator of adiponectin, and enhances SREBP-1c–mediated adiponectin promoter activation. Id3/E47 and E47/SREBP-1c were shown to interact, suggesting that the action of Id3 on SREBP-1c takes place through inhibition of the SREBP-1c interacting partner, E47. As a result, E47 binding to the adiponectin promoter was significantly reduced by the overexpression of Id3 in vitro and conversely, lack of Id3 resulted in increased adiponectin expression in Id3−/− adipose tissue (58). These studies contradict previous studies that Id3 directly binds to SREBP-1c (52). The latter studies instead propose that Id3 regulates SREBP-1c activity indirectly by interacting with E47, thus preventing E47 binding to the SREBP-1c leading to impaired activation of adiponectin promoter and its expression (Figure 3B). Adiponectin plays an important role in glucose metabolism but Id3 deficiency did not appear to affect glucose metabolism or insulin sensitivity (54). Moreover, a failure to expand visceral adipose tissue in Id3−/− mice in response to HFD should lead to ectopic accumulation of lipids in the liver, BAT, kidney and skeletal muscle, leading to insulin resistance. However, such a phenotype was not observed in Id3−/− mice. A possible explanation could be that energy expenditure is increased in Id3−/− mice, leading to increased thermogenesis and/or fatty acid oxidation, resulting in normal insulin sensitivity. However, whether Id3 has any significant role in BAT or fatty acid oxidation is largely unknown. Further in vivo studies are required to understand if Id3 has any specific function in energy expenditure, energy balance, fatty acid oxidation and insulin signaling.
3.4. Id4
A role for Id4 in adipocyte differentiation first emerged when the expression of Id4 mRNA was analyzed during in vitro differentiation of 3T3-L1 preadipocytes. Id4 mRNA level was low in confluent undifferentiated preadipocytes but rapidly increased during differentiation (59). Knockdown of Id4 by shRNA in 3T3-L1 cells resulted in significant reduction in intracellular lipid accumulation with a concomitant decrease in the expression levels of C/EBPα and PPARγ and its downstream target genes such as aP2, Glut4 and LPL, suggesting that adipocyte differentiation is impaired in vitro in the absence of Id4 (60). Expression analysis of Id4 in adult mice and human adipose tissues revealed abundant expression of Id4 in these tissues (59), suggesting a possible in vivo role for Id4 in adipose tissue metabolism. To dissect the specific functions of Id4 in adipose tissue differentiation and metabolism, later studies utilized Id4−/− mice and discovered a reduction in both WAT and BAT mass in Id4−/− mice compared to control mice (60). MEFs isolated from Id4−/− embryos and WAT harvested from Id4−/− mice have diminished expression of C/EBPα and PPARγ and its downstream target genes aP2, Glut4, and LPL, suggesting that adipocyte differentiation and WAT development are impaired in Id4−/− mice. As a result, Id4-deficient mice are leaner compared to wild-type controls, and this lean phenotype is especially more apparent when the mice were fed a HFD (60). Since genetic mouse models such as C/EBPα and PPARγ-deficient mice with impaired adipose tissue development are protected from adipocyte hypertrophy when fed a HFD, the lean phenotype could be attributed to defective adipocyte differentiation in Id4−/− mice. However, it was proposed that the observed lean phenotype could be mainly due to a defect in fatty acid uptake or fat storage in the WAT of Id4−/− mice as they fail to efficiently take up, synthesize and/or store triglycerides (60). However, the molecular mechanism by which Id4 facilitates fatty acid uptake or triglyceride storage in the WAT is completely unknown. Insulin plays an essential role in both fatty acid uptake by adipocytes and adipocyte specific lipogenesis. Adipocyte-derived LPL is essential for efficient fatty acid uptake and storage, and insulin triggers PI3K–mediated induction of LPL activity. LPL hydrolyzes circulating triacylglycerols in lipoproteins into glycerol and fatty acids, whose entry into adipocytes is mediated by fatty acid transporter protein 1 (FATP1), which catalyses the conversion of fatty acids into fatty acyl-CoA. Insulin induces fatty acid uptake in adipocytes by stimulating translocation of FATP1 from intracellular vesicles to the plasma membrane (61). Subsequently, insulin promotes the binding of bHLH-LZ transcription factors, upstream stimulatory factor-1 and 2 (USF1 and USF2), to the FAS promoter. Insulin by signaling through protein phosphatase-1 dephosphorylates and activates DNA dependent protein kinase (DNA-PK), which in turn phosphorylates and activates USFs. USFs directly interact with SREBP-1c, resulting in a highly synergistic transcriptional activation of the FAS promoter, leading to lipogenesis (62). However, it is unclear which component of this fatty acid uptake and lipogenesis pathway is regulated by Id4. One possibility could be that Id4 activates the PI3K/Akt pathway similar to its other family member Id1. Although activation of PI3K/Akt by Id1 was not demonstrated directly in the adipocytes, such an activation mechanism was evident in oesophageal cancer cells (63). In the absence of Id4, impaired activation of the PI3K/Akt pathway might lead to impaired activation of LPL, leading to defective fatty acid uptake and storage. However, it is purely speculative, and further studies are required to test this hypothesis and pinpoint a molecular link between the Id4 and PI3K/Akt pathways.
In contrast to its role in white preadipocyte differentiation, Id4 appears to have quite an opposite function during mesenchymal stem cell (MSC) differentiation. MSCs are the progenitors from which osteoblasts, chondrocytes and adipocytes are derived, and MSC differentiation is a very precisely regulated process (64, 65). Knockdown of Id4 in the stromal cell line ST2 resulted in increased adipogenesis with induced expression of PPARγ and its downstream targets. Subsequently, histological analysis of Id4−/− tibia disclosed a drastic increase in the number of adipocytes in epiphyseal bone marrow of tibia and in the lateral calvaria compared to control mice. Consistent with this observation, PPARγ expression was increased in bone marrow cells of the tibia and femur of Id4−/− mice. These observations suggest that Id4 could function as a critical regulator in the lineage choice of MSCs differentiating into either osteoblasts or adipocytes. Conversely, overexpression of Id4 in ST2 cells partially inhibited adipogenesis with decreased lipid accumulation and increased osteogenesis, suggesting that Id4 alters the MSC differentiation program and suppresses adipogenesis and promotes osteogenesis (66). It appears that Id4 switches the direction of osteoblast and adipocyte differentiation by selectively regulating the transcriptional programing of MSC differentiation. Subsequently, an attempt was made to identify candidate bHLH transcription factors that bind to Id4, and discovered direct binding of Id4 with Hey2 (Hairy and Enhancer of Split-related with YRPW motif 2), a direct target of canonical Notch signaling that is involved in osteogenesis and bone formation. Hey2 forms heterodimers with Hes1, and the Hey2/Hes1 complex binds to the E-box motifs and represses transcription (67, 68). Therefore, a direct interaction of Id4 with Hey2 suggests that Id4 might regulate the transcriptional repression of Hey2/Hes1 during osteogenesis. Consistent with this hypothesis, Id4 reversed the transcriptional repression by Hey2/Hes1 heterodimer in a dose-dependent manner (66). However, the contrasting roles of Id4 in white adipocyte differentiation and MSC differentiation program highlight that additional studies are required to fully understand the transcriptional regulatory functions of Id4 during the adipogenesis and MSC differentiation programs in bone marrow.
4. CONCLUSIONS
All 4 Id proteins (Id1- Id4) are expressed in cultured preadipocytes but their differential expression patterns during adipogenesis are quite different, suggesting unique roles for each of the Id proteins in the regulation of adipocyte differentiation and adipose tissue development. The expression pattern of Id1 during adipocyte differentiation is exactly opposite to the expression of other members of the family, i.e., Id2 and Id4, which are induced during adipogenesis. Lack of either Id2 or Id4 resulted in defective adipogenesis. In contrast, Id1 is targeted for degradation during adipocyte differentiation and lack of Id1 accelerated adipogenesis although overall, it did not cause an abnormal increase in total adipose mass in vivo. Overexpression of Id2 and Id4 enhanced adipocyte differentiation, but the consequence of Id1 overexpression on adipocyte differentiation is inconclusive due to targeted degradation of overexpressed Id1 during adipogenesis. Therefore, Id1 appears to have a distinct function during adipocyte differentiation compared to Id2 or Id4. Overall, the Id proteins appear to have non-overlapping functions during adipocyte differentiation, and it would be interesting to investigate how the Id proteins coordinate with one another during adipogenesis. In this regard, future studies should be directed at generating and analyzing adipose tissue development in compound Id-null mice. In addition to their non-overlapping functions during adipogenesis, Id proteins also appear to have very distinctive functions in adipose tissue metabolism. From the studies conducted thus far, it appears that Id1 is mostly involved in BAT-mediated thermogenesis. In contrast, Id2 appears to regulate fatty acid metabolism, and Id4 controls fatty acid uptake, triglyceride synthesis/storage and also regulates the MSC differentiation program. Id3 also seems to have some overlapping function in fatty acid and adipokine metabolism but its role in adipose tissue vascularization is unique to other family members. Although studies performed in all 4 Id null mice provided some basic understanding of each of the four Id proteins in adipose tissue development and metabolism, it appears that the current available information is not quite sufficient to establish a clear connection between different Id proteins and their specific regulatory mechanisms in adipose tissue development and metabolism. Specific deletion of the Id proteins in adipose tissues by crossing Id conditional alleles with aP2cre mice will possibly provide more in-depth information regarding the involvement of individual members of the Id family of proteins in adipose tissue-specific metabolic pathways.
ACKNOWLEDGMENTS
The authors thank Dr. Rhea-Beth Markowitz for critically reviewing the manuscript. This research is supported by the National Cancer Institute (NCI) of the National Institutes of Health (K22CA168828).
Abbreviations
- ADD1
Adipocyte determination and differentiation factor 1
- Akt
AKT1 kinase
- aP2
Fatty acid binding protein
- APC
Adenomatous polyposis coli
- ARRDC3
Arrestin domain containing 3
- bHLH
basic Helix-loop-helix
- bHLH-LZ
basic helix-loop-helix leucine zipper
- BAT
Brown adipose tissue
- BMP
Bone morphogenetic protein
- c-Myc
Myelocytomatosis oncogene
- C/EBP
CCAAT/enhancer binding protein
- CD36
Thrombospondin receptor
- CDK
Cyclin dependent kinase
- Cidea
Cell death-inducing DNA fragmentation factor, alpha subunit-like effector A
- Cre
Cyclization recombinase
- DNA-PK
DNA dependent protein kinase
- DP
E2F dimerization partner
- E2F
E2F transcription factor
- Ebf2
Early B cell factor 2
- Ets2
Avian erythroblastosis virus E26 (v-ets) oncogene homolog 2
- FAS
Fatty acid synthase
- FATP1
Fatty acid transporter protein 1
- FOXC2
Fork-head box C2
- GATA
GATA binding protein
- Glut4
Glucose transporter type 4
- GSK3
Glycogen synthase kinase 3
- Hes 1
Hairy and enhancer of split 1
- Hey 2
Hairy and enhancer of split-related with YRPW motif 2
- HFD
High fat diet
- HLH
Helix-loop-helix
- Id
Inhibitor of DNA binding
- KLF
Kruppel-like factor
- LPL
Lipoprotein lipase
- LSD1 demethylase
Lysine specific demethylase 1
- LXR
Liver-X-receptor
- MEFs
Mouse embryonic fibroblasts
- MEK
Mitogen activated protein kinase kinase
- MSCs
Mesenchymal stem cells
- Myf-5
Myogenic factor 5
- MyoD
Myogenic differentiation protein
- NAFLD
Non-alcoholic fatty liver disease
- PAX
Paired box gene
- PGC1α
Peroxisome proliferator activated receptor gamma coactivator 1 alpha
- PI3K
Phosphatidylinositol 3-kinase
- PPAR
Peroxisome proliferator-activated receptor
- pRB
Retinoblastoma protein
- PRDM16
PR domain containing 16
- Ras
Rat sarcoma viral oncogene homolog
- Raf
Raf kinase
- RARs
Retinoic acid receptors
- RER
Respiratory exchange ratio
- siRNA
Small interfering RNA
- Smurf2
SMAD specific E3 ubiquitin protein ligase 2
- ST2 cells
Stromal cell line
- STAT
Signal-transducer and activator of transcription protein
- SVF
Stromal vascular fraction
- SREBP-1c
Sterol regulatory element-binding protein 1c
- SRC2
Src tyrosine kinase 2
- TCF/LEF
T-cell factor/lymphoid enhancer factor
- TRα1
Thyroid hormone receptor alpha
- UCP1
Uncoupling protein 1
- USF1
Upstream stimulatory factor 1
- USF2
Upstream stimulatory factor 2
- VEGF
Vascular endothelial growth factor
- WAT
White adipose tissue
- Wnt
Wingless
- WRE
Wnt response element
Footnotes
The authors declare no competing financial conflicts of interest.
REFERENCES
- 1.Malik VS, Willett WC, Hu FB. Global obesity: trends, risk factors and policy implications. Nature reviews Endocrinology. 2013;9:13–27. doi: 10.1038/nrendo.2012.199. [DOI] [PubMed] [Google Scholar]
- 2.McAllister EJ, Dhurandhar NV, Keith SW, Aronne LJ, Barger J, Baskin M, Benca RM, Biggio J, Boggiano MM, Eisenmann JC, Elobeid M, Fontaine KR, Gluckman P, Hanlon EC, Katzmarzyk P, Pietrobelli A, Redden DT, Ruden DM, Wang C, Waterland RA, Wright SM, Allison DB. Ten putative contributors to the obesity epidemic. Critical reviews in food science and nutrition. 2009;49:868–913. doi: 10.1080/10408390903372599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li C, Hsieh MC, Chang SJ. Metabolic syndrome, diabetes, and hyperuricemia. Current opinion in rheumatology. 2013;25:210–216. doi: 10.1097/BOR.0b013e32835d951e. [DOI] [PubMed] [Google Scholar]
- 4.Xu WL, Atti AR, Gatz M, Pedersen NL, Johansson B, Fratiglioni L. Midlife overweight and obesity increase late-life dementia risk: a population-based twin study. Neurology. 2011;76:1568–1574. doi: 10.1212/WNL.0b013e3182190d09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirza MS. Obesity, Visceral Fat, and NAFLD: Querying the Role of Adipokines in the Progression of Nonalcoholic Fatty Liver Disease. ISRN gastroenterology. 2011;2011:592404. doi: 10.5402/2011/592404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirier P, Eckel RH. Obesity and cardiovascular disease. Current atherosclerosis reports. 2002;4:448–453. doi: 10.1007/s11883-002-0049-8. [DOI] [PubMed] [Google Scholar]
- 7.Bardou M, Barkun AN, Martel M. Obesity and colorectal cancer. Gut. 2013;62:933–947. doi: 10.1136/gutjnl-2013-304701. [DOI] [PubMed] [Google Scholar]
- 8.Vucenik I, Stains JP. Obesity and cancer risk: evidence, mechanisms, and recommendations. Annals of the New York Academy of Sciences. 2012;1271:37–43. doi: 10.1111/j.1749-6632.2012.06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hursting SD, Dunlap SM. Obesity, metabolic dysregulation, and cancer: a growing concern and an inflammatory (and microenvironmental) issue. Annals of the New York Academy of Sciences. 2012;1271:82–87. doi: 10.1111/j.1749-6632.2012.06737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hursting SD. Obesity, energy balance, and cancer: a mechanistic perspective. Cancer treatment and research. 2014;159:21–33. doi: 10.1007/978-3-642-38007-5_2. [DOI] [PubMed] [Google Scholar]
- 11.Klaus S. Functional differentiation of white and brown adipocytes. BioEssays : news and reviews in molecular, cellular and developmental biology. 1997;19:215–223. doi: 10.1002/bies.950190307. [DOI] [PubMed] [Google Scholar]
- 12.Spiegelman BM, Flier JS. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 13.Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, Fujimoto Y, Shiota M, Kesterson RA, Kahn BB, Magnuson MA. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B, Bick RJ, Darlington GJ. C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12532–12537. doi: 10.1073/pnas.211416898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes & development. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 16.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 17.Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 18.Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell metabolism. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sellayah D, Bharaj P, Sikder D. Orexin is required for brown adipose tissue development, differentiation, and function. Cell metabolism. 2011;14:478–490. doi: 10.1016/j.cmet.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, Zhang Y, Dobrin R, Cohen DE, Larsen PR, Zavacki AM, Fong LG, Young SG, Lee RT. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell metabolism. 2011;14:671–683. doi: 10.1016/j.cmet.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, Lin SC, Hong W, Li P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nature genetics. 2003;35:49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
- 22.Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan D, Fujimoto M, Lopes A, Wang YX. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell. 2009;137:73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function--of mice and men. Genes & development. 2009;23:788–797. doi: 10.1101/gad.1779209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell metabolism. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz TJ, Huang P, Huang TL, Xue R, McDougall LE, Townsend KL, Cypess AM, Mishina Y, Gussoni E, Tseng YH. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 28.Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 29.Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20:8317–8325. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
- 30.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 31.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nature reviews Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 32.Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nature reviews. Immunology. 2001;1:193–199. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- 33.Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. Journal of cell science. 2000;113(Pt 22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 34.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends in cell biology. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 35.Satyanarayana A, Klarmann KD, Gavrilova O, Keller JR. Ablation of the transcriptional regulator Id1 enhances energy expenditure, increases insulin sensitivity, and protects against age and diet induced insulin resistance, and hepatosteatosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:309–323. doi: 10.1096/fj.11-190892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7812–7816. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 38.Zheng W, Wang H, Xue L, Zhang Z, Tong T. Regulation of cellular senescence and p16(INK4a) expression by Id1 and E47 proteins in human diploid fibroblast. The Journal of biological chemistry. 2004;279:31524–31532. doi: 10.1074/jbc.M400365200. [DOI] [PubMed] [Google Scholar]
- 39.Classon M, Kennedy BK, Mulloy R, Harlow E. Opposing roles of pRB and p107 in adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10826–10831. doi: 10.1073/pnas.190343597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L, Nerlov C. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–258. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- 41.Farmer SR. Transcriptional control of adipocyte formation. Cell metabolism. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trausch-Azar JS, Lingbeck J, Ciechanover A, Schwartz AL. Ubiquitin-Proteasome-mediated degradation of Id1 is modulated by MyoD. The Journal of biological chemistry. 2004;279:32614–32619. doi: 10.1074/jbc.M403794200. [DOI] [PubMed] [Google Scholar]
- 43.Kong Y, Cui H, Zhang H. Smurf2-mediated ubiquitination and degradation of Id1 regulates p16 expression during senescence. Aging cell. 2011;10:1038–1046. doi: 10.1111/j.1474-9726.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park KW, Waki H, Villanueva CJ, Monticelli LA, Hong C, Kang S, MacDougald OA, Goldrath AW, Tontonoz P. Inhibitor of DNA binding 2 is a small molecule-inducible modulator of peroxisome proliferator-activated receptor-gamma expression and adipocyte differentiation. Molecular endocrinology. 2008;22:2038–2048. doi: 10.1210/me.2007-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karaya K, Mori S, Kimoto H, Shima Y, Tsuji Y, Kurooka H, Akira S, Yokota Y. Regulation of Id2 expression by CCAAT/enhancer binding protein beta. Nucleic acids research. 2005;33:1924–1934. doi: 10.1093/nar/gki339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual review of cell and developmental biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 47.Modarresi R, Xiang Z, Yin M, Laurence J. WNT/beta-catenin signaling is involved in regulation of osteoclast differentiation by human immunodeficiency virus protease inhibitor ritonavir: relationship to human immunodeficiency virus-linked bone mineral loss. The American journal of pathology. 2009;174:123–135. doi: 10.2353/ajpath.2009.080484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Memezawa A, Takada I, Takeyama K, Igarashi M, Ito S, Aiba S, Kato S, Kouzmenko AP. Id2 gene-targeted crosstalk between Wnt and retinoid signaling regulates proliferation in human keratinocytes. Oncogene. 2007;26:5038–5045. doi: 10.1038/sj.onc.1210320. [DOI] [PubMed] [Google Scholar]
- 49.Farmer SR. Regulation of PPARgamma activity during adipogenesis. International journal of obesity. 2005;29(Suppl 1):S13–S16. doi: 10.1038/sj.ijo.0802907. [DOI] [PubMed] [Google Scholar]
- 50.Ji M, Li H, Suh HC, Klarmann KD, Yokota Y, Keller JR. Id2 intrinsically regulates lymphoid and erythroid development via interaction with different target proteins. Blood. 2008;112:1068–1077. doi: 10.1182/blood-2008-01-133504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathew D, Zhou P, Pywell CM, van der Veen DR, Shao J, Xi Y, Bonar NA, Hummel AD, Chapman S, Leevy WM, Duffield GE. Ablation of the ID2 gene results in altered circadian feeding behavior, and sex-specific enhancement of insulin sensitivity and elevated glucose uptake in skeletal muscle and brown adipose tissue. PloS one. 2013;8:e73064. doi: 10.1371/journal.pone.0073064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moldes M, Boizard M, Liepvre XL, Feve B, Dugail I, Pairault J. Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element-binding protein-1c (ADD1/SREBP-1c) trans-factors for the regulation of fatty acid synthase promoter in adipocytes. The Biochemical journal. 1999;3(344 Pt):873–880. [PMC free article] [PubMed] [Google Scholar]
- 53.Moldes M, Lasnier F, Feve B, Pairault J, Djian P. Id3 prevents differentiation of preadipose cells. Molecular and cellular biology. 1997;17:1796–1804. doi: 10.1128/mcb.17.4.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cutchins A, Harmon DB, Kirby JL, Doran AC, Oldham SN, Skaflen M, Klibanov AL, Meller N, Keller SR, Garmey J, McNamara CA. Inhibitor of differentiation-3 mediates high fat diet-induced visceral fat expansion. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:317–324. doi: 10.1161/ATVBAHA.111.234856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 56.Bryan BA, Walshe TE, Mitchell DC, Havumaki JS, Saint-Geniez M, Maharaj AS, Maldonado AE, D'Amore PA. Coordinated vascular endothelial growth factor expression and signaling during skeletal myogenic differentiation. Molecular biology of the cell. 2008;19:994–1006. doi: 10.1091/mbc.E07-09-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Doran AC, Meller N, Cutchins A, Deliri H, Slayton RP, Oldham SN, Kim JB, Keller SR, McNamara CA. The helix-loop-helix factors Id3 and E47 are novel regulators of adiponectin. Circulation research. 2008;103:624–634. doi: 10.1161/CIRCRESAHA.108.175893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen H, Weng YC, Schatteman GC, Sanders L, Christy RJ, Christy BA. Expression of the dominant-negative regulator Id4 is induced during adipocyte differentiation. Biochemical and biophysical research communications. 1999;256:614–619. doi: 10.1006/bbrc.1999.0386. [DOI] [PubMed] [Google Scholar]
- 60.Murad JM, Place CS, Ran C, Hekmatyar SK, Watson NP, Kauppinen RA, Israel MA. Inhibitor of DNA binding 4 (ID4) regulation of adipocyte differentiation and adipose tissue formation in mice. The Journal of biological chemistry. 2010;285:24164–24173. doi: 10.1074/jbc.M110.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czech MP, Tencerova M, Pedersen DJ, Aouadi M. Insulin signalling mechanisms for triacylglycerol storage. Diabetologia. 2013;56:949–964. doi: 10.1007/s00125-013-2869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffin MJ, Sul HS. Insulin regulation of fatty acid synthase gene transcription: roles of USF and SREBP-1c. IUBMB life. 2004;56:595–600. doi: 10.1080/15216540400022474. [DOI] [PubMed] [Google Scholar]
- 63.Li B, Cheung PY, Wang X, Tsao SW, Ling MT, Wong YC, Cheung AL. Id-1 activation of PI3K/Akt/NFkappaB signaling pathway and its significance in promoting survival of esophageal cancer cells. Carcinogenesis. 2007;28:2313–2320. doi: 10.1093/carcin/bgm152. [DOI] [PubMed] [Google Scholar]
- 64.Muruganandan S, Roman AA, Sinal CJ. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: cross talk with the osteoblastogenic program. Cellular and molecular life sciences : CMLS. 2009;66:236–253. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dennis JE, Charbord P. Origin and differentiation of human and murine stroma. Stem cells. 2002;20:205–214. doi: 10.1634/stemcells.20-3-205. [DOI] [PubMed] [Google Scholar]
- 66.Tokuzawa Y, Yagi K, Yamashita Y, Nakachi Y, Nikaido I, Bono H, Ninomiya Y, Kanesaki-Yatsuka Y, Akita M, Motegi H, Wakana S, Noda T, Sablitzky F, Arai S, Kurokawa R, Fukuda T, Katagiri T, Schonbach C, Suda T, Mizuno Y, Okazaki Y. Id4, a new candidate gene for senile osteoporosis, acts as a molecular switch promoting osteoblast differentiation. PLoS genetics. 2010;6:e1001019. doi: 10.1371/journal.pgen.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Molecular and cellular biology. 2001;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zanotti S, Canalis E. Hairy and Enhancer of Split-related with YRPW motif (HEY)2 regulates bone remodeling in mice. The Journal of biological chemistry. 2013;288:21547–21557. doi: 10.1074/jbc.M113.489435. [DOI] [PMC free article] [PubMed] [Google Scholar]