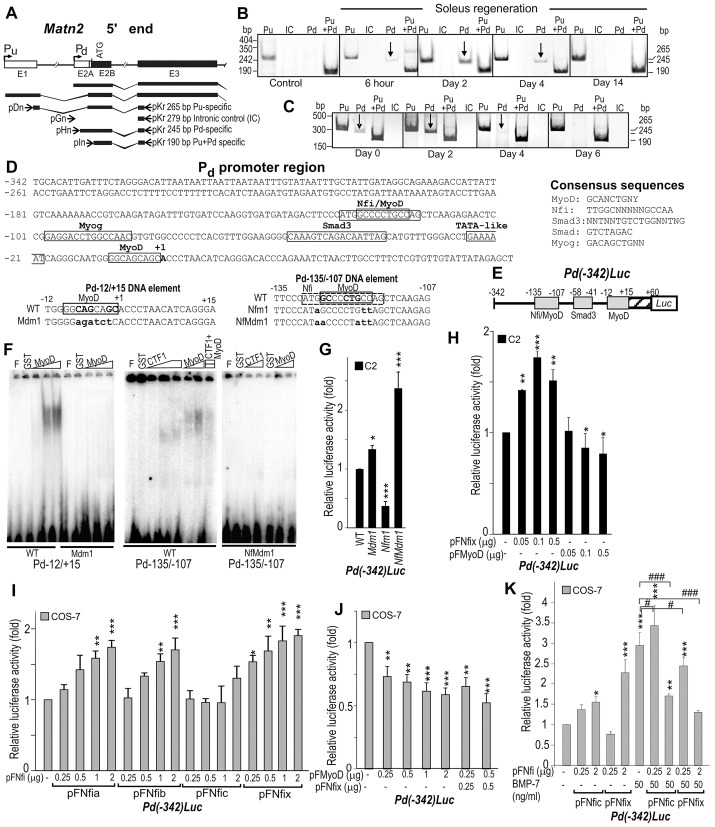
Fig. 6.
Regulation of the Matn2 promoter by MyoD and Nfi. (A) Schematic illustrating the exons E1–E3, the two alternative promoters and the Matn2 transcripts. Primer pairs specific for Pu and Pd and for intronic control (IC) are depicted (arrows), and have been described previously (Mátés et al., 2002). Semi-quantitative RT-PCR analysis using Pu- and Pd-specific and control primer pairs during soleus regeneration (B) and C2 myoblast differentiation (C). (D) Nucleotide sequences showing the putative factor-binding sites in the Pd region and in the wild-type (WT) and mutant versions of DNA elements Pd−12/+15 and Pd−135/−107. Mutated nucleotides are shown in lower case. (E) Structure of the Pd-luciferase fusion construct. (F) The binding of purified GST (glutathione S-transferase), GST–MyoD and GST–CTF1 (NFIC isoform) to the wild-type and mutant Pd−12/+15 and Pd−135/−107 DNA elements was determined in EMSA. F, free probe. (G) The effect of the NFI and MyoD motif mutations shown in D on the Pd activity of Pd(−342)Luc in C2 myoblasts. (H–K) The effect of forced expression of NFI proteins and MyoD from the indicated amounts of expression plasmids (pFNfi and pFMyoD) on the luciferase activity of Pd(−342)Luc in C2 myoblasts and COS-7 cells. In COS-7 cells, NFI proteins can increase Pd activity (I), whereas MyoD alone or upon coexpression with Nfix can repress it (J). Bmp7 can increase Pd activity and its dose-dependent activation by Nfic and Nfix (K). Data represent the mean±s.e.m.; *P<0.05, **P<0.01, ***P<0.001 [relative to Pd(−342)Luc], #P<0.05, ###P<0.001 (as indicated).