Abstract
Synchronous cultures are often indispensable for studying meiosis. Here, we present an optimized protocol for induction of synchronous meiosis in the fission yeast Schizosaccharomyces pombe. Chemical inactivation of an ATP analog-sensitive form of the Pat1 kinase (pat1-as2) by adding the ATP-analog 1-NM-PP1 in G1-arrested cells allows induction of synchronous meiosis at optimal temperature (25 °C). Importantly, this protocol eliminates detrimental effects of elevated temperature (34 °C) which is required to inactivate the commonly used temperature-sensitive Pat1 kinase mutant (pat1-114). Addition of the mat-Pc gene to a mat1-M strain further improves chromosome segregation and spore viability. Thus, our protocol offers highly synchronous meiosis at optimal temperature with most characteristics similar to those of wild-type meiosis. The synchronization protocol can be completed in 5 days.
Introduction
Sexual reproduction in eukaryotes depends on meiosis, the process which produces haploid gametes (e.g. egg and sperm cells in animals, ovules and pollen in flowering plants or spores in fungi) from diploid precursor cells. Two rounds of chromosome segregation after only a single round of DNA replication enable the reduction of chromosome number. While the second meiotic division is similar to mitosis in that the centromeres of sister chromatids segregate to opposite poles, the first meiotic division is fundamentally different and ensures segregation of centromeres of homologous chromosomes (homologs), usually after recombination (crossing over) between homologs1,2.
The fission yeast Schizosaccharomyces pombe is an excellent model organism for studying molecular mechanisms governing meiosis as it is amenable to both genetic and cell biological techniques, and highly synchronous meiosis can be induced3–7.
Moreover, fission yeast is in some ways more similar to metazoans than is the other popular yeast model, Saccharomyces cerevisiae (budding yeast): S. pombe has large, complex centromeres; it harbors histone H3 Lys9 methylation and chromodomain proteins that, with RNA interference (RNAi), regulate gene expression, transposon mobility, and meiotic recombination8 ; and its genes have abundant introns9. Since many of the processes that ensure the reductional nature of chromosome segregation during meiosis are conserved in evolution, this makes the fission yeast an excellent model for studying chromosome biology during meiosis, although it seems to lack crossover interference4.
Development of the protocol and comparison with other methods
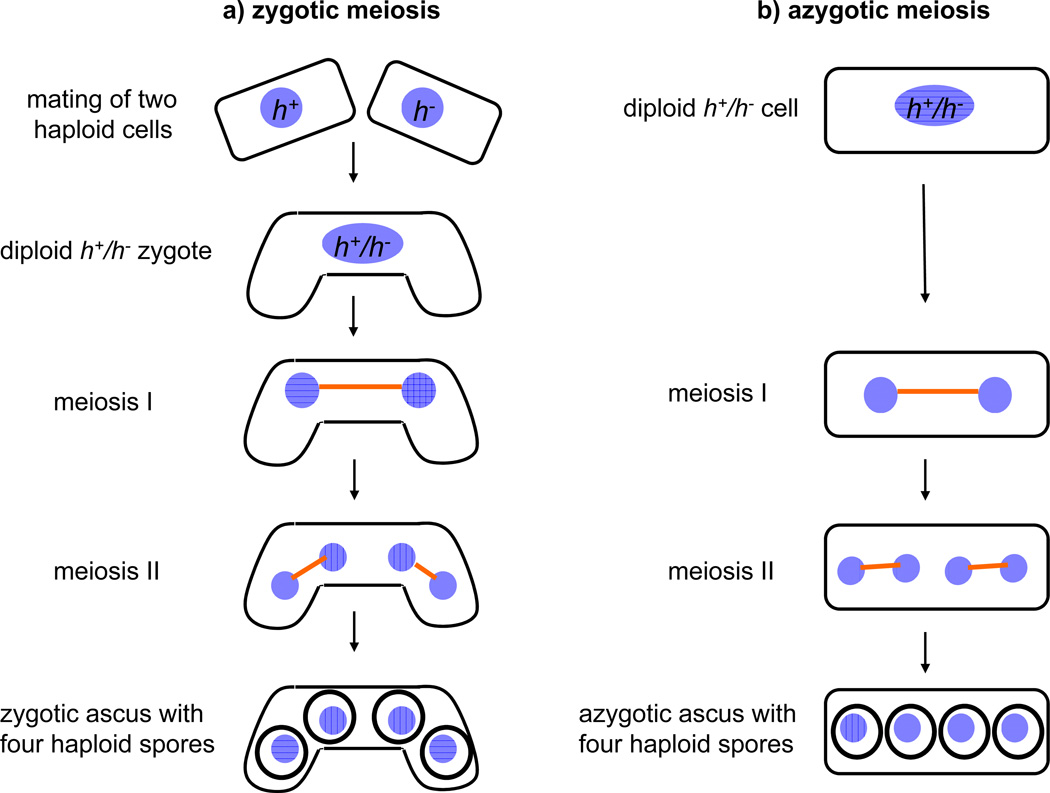
Fission yeast cells can proliferate as haploids, which have one of two mating types, h+ (P) and h− (M). Mating type is determined by the mat1 locus, which contains two genes (i-type and c-type) that differ in h+ (P) and h− (M). Upon nitrogen starvation, haploid cells of either mating type (h+ or h−) arrest in G1 phase. Subsequently, cells of opposite mating type mate to form diploid zygotes, which undergo meiosis (this is called zygotic meiosis) and generate asci with four haploid spores10 (Figure 1). This process is very efficient but asynchronous. For biochemical analyses, synchronous meiotic cultures are often indispensable. There are two major methods that allow induction of synchronous meiosis in S. pombe: 1) inactivation of a conditional form of the Pat1 kinase, and 2) using a diploid strain heterozygous at the mating-type locus.
Figure 1. Zygotic and azygotic meiosis in the fission yeast S. pombe.
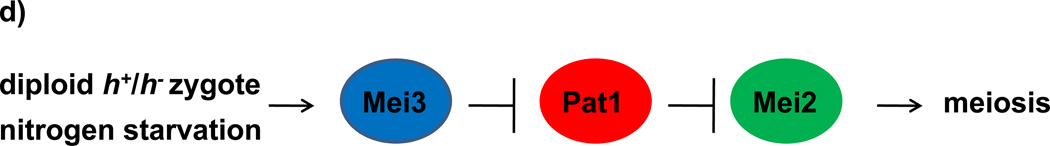
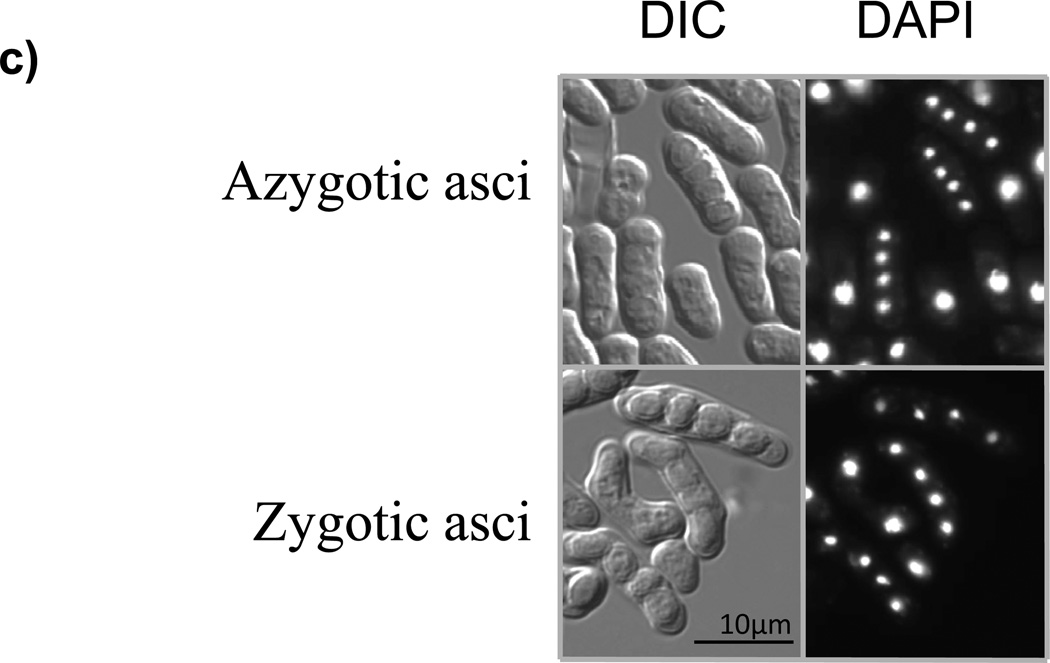
(a) Scheme of zygotic meiosis. Haploid cells of the opposite mating type (h+ and h−) mate to form diploid zygotes, which undergo two meiotic divisions (this is called zygotic meiosis) and generate asci, typically curved, with four haploid spores. (b) Scheme of azygotic meiosis. Diploid cells heterozygous at the mating type locus (h+/h−) enter meiosis and generate azygotic asci, typically straight, with four haploid spores. (c) DIC (differential interference contrast) images and DAPI staining of azygotic asci from wild-type diploid h+ /h− strain (JG16539) and zygotic asci from wild-type h90 strain (JG11355). (d) Regulation of initiation of meiosis in the fission yeast S. pombe. Mei3 is expressed upon activation of the mating pheromone signaling cascade (MAPK) and starvation for nitrogen. Mei3 inhibits Pat1 kinase, and hence liberates Mei2 from its inhibitory phosphorylation. Additional factors, including the Ste11 transcription factor, are also important.
A temperature-sensitive allele (pat1-114) of the Pat1 (Ran1) protein kinase allows induction of synchronous meiosis in S. pombe independently of ploidy and most nutritional signals11–14. Pat1 kinase inhibits meiosis by negatively regulating an RNA-binding protein Mei2 which is the major target of Pat1 during mitotic growth15,16 (Figure 2). Cells harboring pat1-114 mutation can be induced to undergo meiosis in a timely and predictable manner by shifting nitrogen-starved cultures from a permissive (25 °C) to restrictive temperature (34 °C)6,11–14. However, pat1-114-induced meiosis differs from wild-type meiosis in some aspects, such as chromosome segregation6,11–15,17–19. While in wild-type cells, sister centromeres nearly always segregate to the same pole in anaphase I, in meiosis induced by inactivation of Pat1- 114 or Pat1-as2 by elevated temperature (34 °C), sister centromeres segregate to the same pole very inefficiently in anaphase I cells (Table 1)17,20. We speculated that some of these abnormalities might be due to the higher temperature needed to inactivate the Pat1 kinase. Therefore, we have applied a chemical-genetics strategy to develop an ATP analog-sensitive form of Pat1 [Pat1(L95A), designated pat1-as2)] which can be used to generate synchronous meiotic cultures at physiological temperature20,21 (Figure 2a, Figure 2b). A similar approach has been successfully developed in E. Hidalgo and J. Ayte's laboratory using the pat1-as1 (Pat1(L95G)) allele18. The pat1-as1 allele developed in our laboratory20 is not fully functional at 25 °C (our unpublished data), therefore we have used pat1-as2. In pat1-as2- induced meiosis, chromosomes segregate with higher fidelity than in pat1-114 meiosis. Moreover, addition of the mat-Pc gene improves the fidelity of chromosome segregation and spore viability to nearly the level of meiosis in wild-type cells (Table 1)17,20. Our recent study shows that many features of meiotic recombination in pat1-as2 cells at 25 °C are the same as those in the pat1-114 mutant at 34 °C; however, some features of two intermediates – DNA double strand breaks and joint DNA molecules at certain hotspots – are different at the two temperatures. Interestingly, a novel species, perhaps arising from invasion by only one end of broken DNA, is more readily observed at 25 °C (Hyppa et al., in press). In addition, pat1-as2-induced meiosis eliminates the effect of heat shock and consequent stress responses on meiosis18. Thus, we conclude that pat1-as2 mat-Pc cells offer highly synchronous meiosis with most tested properties similar to those of wild-type meiosis. This improved synchronization protocol is therefore a useful tool for studying, among other things, chromosome segregation and recombination in fission yeast meiosis22–26.
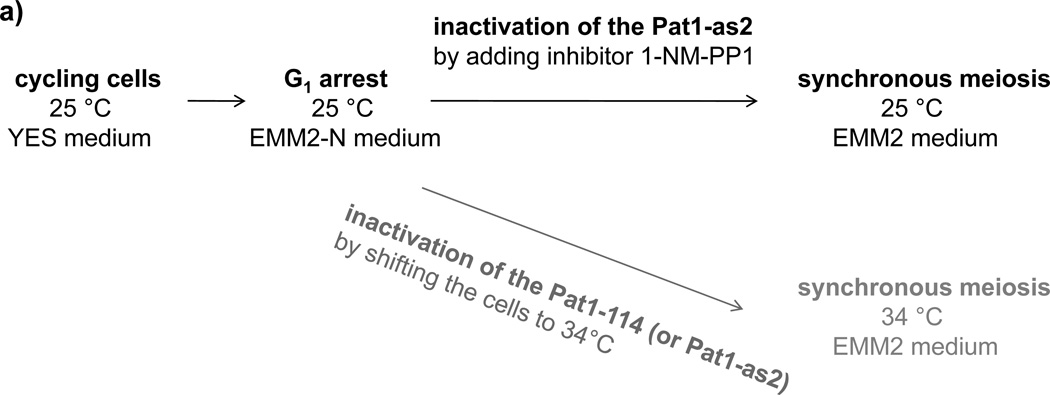
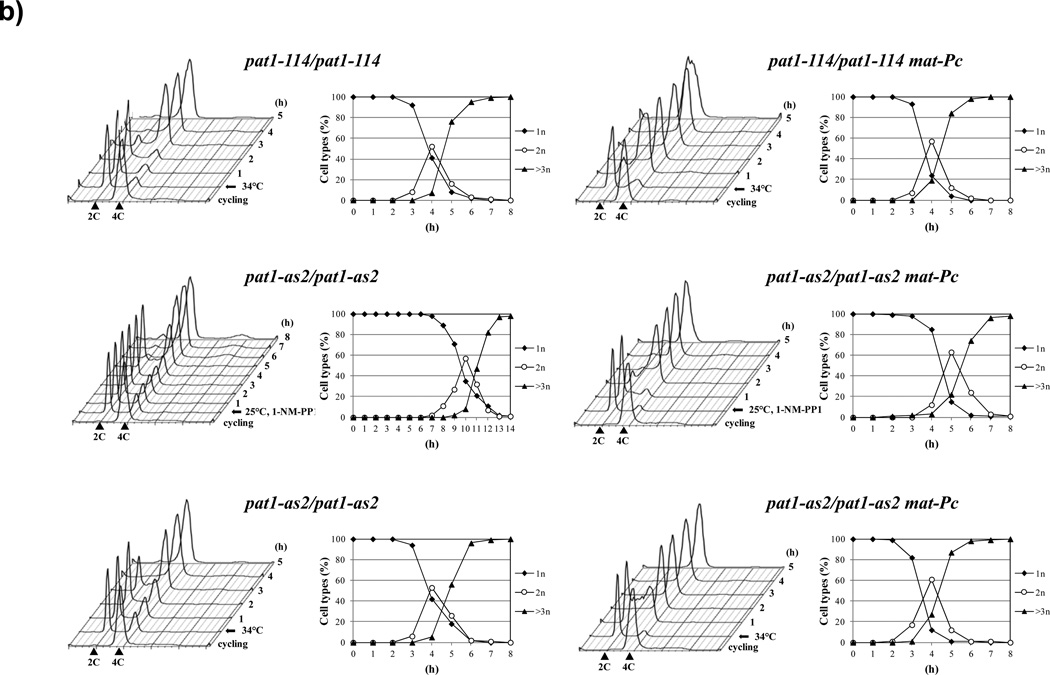
Figure 2. Synchronous meiosis induced by inactivation of Pat1.
(a) Flowchart of the pat1 mutant-induced synchronous meiosis. Cells carrying the pat1-as2 or pat1-114 allele are grown in liquid YES medium to mid-log phase. The cells are collected by centrifugation, resuspended in liquid EMM2 medium lacking a nitrogen source (EMM2-NH4Cl) and incubated at 25 °C to arrest cells in G1 phase. Subsequently, the cells are resuspended in fresh EMM2 medium containing a nitrogen source and synchronous meiosis is induced in pat1-as2 cells by adding the inhibitor 1-NM-PP1. Alternatively, Pat1-as2 (or Pat1-114) can be inactivated by shifting the cells to non-permissive temperature 34 °C. (b) Progression of meiosis in diploid strains pat1-114/pat1-114 (JG12209), pat1-as2/pat1-as2 (JG15620), pat1- 114/pat1-114 mat-Pc (JG16328) and pat1-as2/pat1-as2 mat-Pc (JG16113). Cells were cultured to mid-log phase in YES-Ade medium, transferred to EMM2-NH4Cl medium for 16 h at 25 °C (pat1-114 and pat1-as2) or for 7 h at 25 °C (pat1-114 mat-Pc and pat1-as2 mat- Pc) to synchronize cells in G1, transferred to EMM2 medium, and incubated at 34 °C or kept at 25 °C with addition of 25 µM 1-NM-PP 1 to inactivate the Pat1-as2 kinase. Progression of meiosis was monitored by flow cytometry for DNA content and by fluorescence microscopy for number of nuclei per cell in samples collected at the indicated time-points after temperature-shift or addition of 1-NM-PP 120.
Table 1.
Analysis of spore viability and segregation of sister centromeres during meiosis I21.
| Strain |
mat1 alleles |
Ectopic mat-Pc |
P-factor addition |
Mutation | Induction condition |
Spore viability |
Reductional anaphase I |
|---|---|---|---|---|---|---|---|
| JG12226 × JG15458 | P/M | - | - | - | 25°C | 93% | 99% |
| JG16022 | M/M | - | - | pat1-as2 | 25°C 1-NM-PP1 | 56% | 44% |
| JG16022 | M/M | - | - | pat1-as2 | 34°C | 32% | 30% |
| JG16022 | M/M | - | P-factor | pat1-as2 | 25°C 1-NM-PP1 | 81% | 90% |
| JG16022 | M/M | - | P-factor | pat1-as2 | 34°C | 74% | 85% |
| JG16113 | M/M | mat-Pc | - | pat1-as2 | 25°C 1-NM-PP1 | 85% | 96% |
| JG16113 | M/M | mat-Pc | - | pat1-as2 | 34°C | 78% | 93% |
Diploid S. pombe cells homozygous at mat1 can be obtained by fusing protoplasts of h+ or hhaploids with complementing auxotrophies27. The ade6-216 and ade6-210 alleles, which manifest intragenic complementation, are often used because recombination between these two markers, which with appropriate chromosome segregation can produce a prototrophic haploid, is very rare. Diploids homozygous at mat1, but otherwise wild-type, do not, however, undergo meiosis. Diploid S. pombe cells heterozygous at the mating type locus can be obtained by crossing h+ and h− haploids with complementary growth requirements (e.g. ade6-M216 and ade6-M210) followed by transfer of zygotes from nitrogen-free medium to growth medium before commitment to meiosis. Only diploid cells will grow in the absence of adenine due to intragenic complementation between the ade6-M216 and ade6-M210 alleles. However, using diploid cells heterozygous at the mating type locus is problematic because they enter meiosis upon reaching stationary phase and diploidy is lost. Meiosis can be induced by shifting exponentially growing diploid S. pombe cells that are heterozygous at mat1 from growth medium to sporulation medium6,28. These cells will undergo successful meiosis (this is called azygotic meiosis) (Figure 1) with some degree of synchrony but not enough for many studies6,29,30.
Other methods to induce meiosis in the fission yeast include overexpression of Mei331, expression of a non-phosphorylatable form of Mei2 (Mei2-SATA)16 and ectopic expression of a constitutively active form of Byr132. All these methods allow induction of meiosis in haploid or diploid cells; however, the degree of synchrony has not been investigated in detail. In addition, a novel temperature-sensitive mei4 allele (mei4-N136A) can be used to synchronize the meiotic cell cycle from meiosis I onwards19. Finally, an optimized protocol for synchronous meiosis at 34 °C using diploid h−/h− pat1-114/pat1-114 cells ectopically expressing mat-Pc has been developed33. In our opinion, this protocol which combines pat1- 114 together with ectopically expressed mat-Pc is the best alternative to our pat1-as2 mat-Pc protocol, although it cannot be carried out at optimal temperature.
Experimental Design
To induce synchronous meiosis, haploid pat1-as2 or diploid pat1-as2/pat1-as2 strains are grown in YE+5S (pat1-as2) or YE+4S-Ade (pat1-as2/pat1-as2) liquid medium to mid-log phase at 25 °C. The cells are collected by centrifugation, resuspended in EMM2-NH4Cl medium and incubated at 25°C to arrest cells in G1. The cells are resuspended in fresh EMM2 medium containing a nitrogen source (NH4Cl) and induced into meiosis by adding the ATPanalog 1-NM-PP1 at 25 °C (Figure 2a). To induce the mating pheromone response in pat1-as2 cells, mat-Pc is integrated into the genome or, alternatively, synthetic P-factor is added to the EMM2 medium. To monitor the progression of meiosis, aliquots of the culture are collected at desired time-points, and meiotic progression is monitored by fluorescence microscopy for nuclear divisions and by flow cytometry for DNA synthesis. When using pat1-as2 strain in combination with another mutation (e.g. pat1-as2 rec8Δ), always use the corresponding wild-type strain as a control (in this case pat1-as2 rec8+); additional controls could include pat1+, to test for “off-target” effects of the inhibitor, and no inhibitor, to test for specificity of meiotic induction.
Limitations
Although most of the pat1-as2 cells undergo meiosis upon adding the inhibitor, there is a small fraction (usually less than 5%) of cells that fail to undergo meiosis.
The inhibitor 1-NM-PP1 is expensive. Given that using analog-sensitive kinase alleles is becoming more popular, we expect that the price will drop in the near future. Alternatively, it may be possible to increase the sensitivity of pat1-as2 cells to 1-NM-PP1 inhibitor by introducing additional mutations in the ATP binding pocket34,35 or by using a mutant strain hyper-sensitive to chemical inhibitors in which the key transcription factors and drug-efflux transporters responsible for multidrug resistance have been mutated36.
In some experiments, it may be desirable to combine pat1-as2 with a temperature-sensitive allele of another essential protein. As the pat1-as2 allele created in our lab is also temperature-sensitive (Figure 3a), some experiments may be difficult to perform. Interestingly, the pat1-as1 allele created in the laboratory of E. Hidalgo and J. Ayte provides synchronous meiosis, but this allele is not temperature-sensitive18 (please note that the pat1- as1 allele created in our laboratory is temperature-sensitive20). In addition, strains carrying suppressor mutations, either intragenic or extragenic, can be isolated by growing pat1-as2 cells at 32 °C (Figure 3a)11,37 ; some of these suppressor strains maintain their sensitivity to 1- NM-PP1 (our unpublished data).
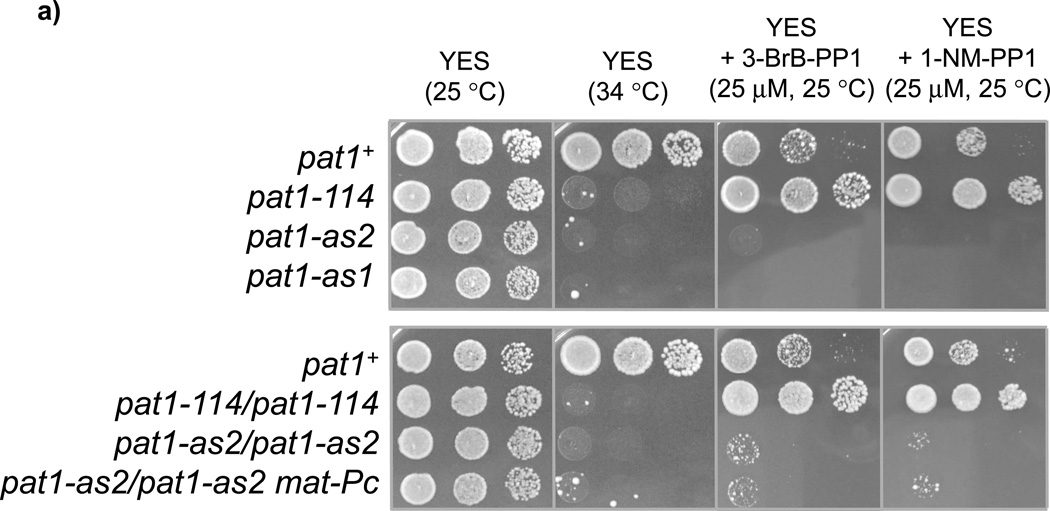
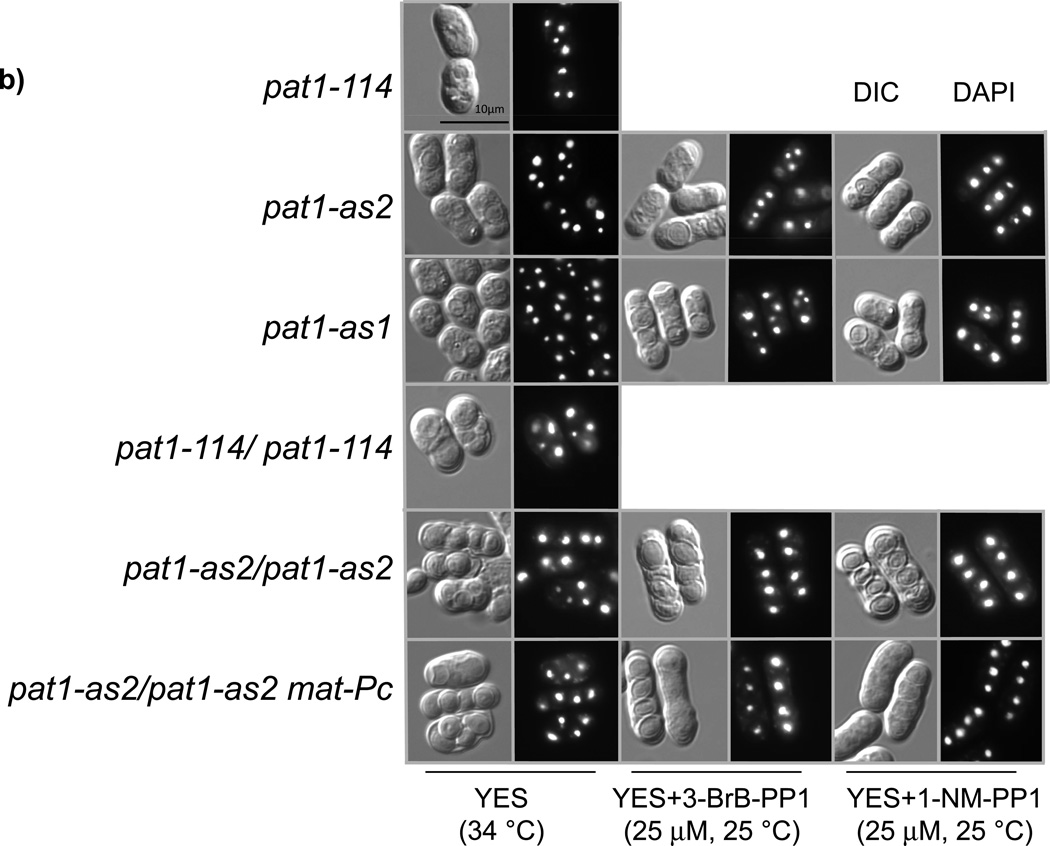
Figure 3. Sensitivity of cells expressing Pat1-as2 to ATP-analogs.
(a) Serial dilutions of wild-type cells (pat1+) (JG15458) or pat1-114 (JG11322), pat1-as1 (JG15404) or pat1-as2 (JG15403) cells as well as diploid cells pat1-114/pat1-114 (JG15710), pat1-as2/pat1-as2 (JG16022) and pat1-as2/pat1-as2 matPc (JG16113) were spotted on YES plates containing or lacking the indicated ATP-analogs (25 µM 3-BrB-PP1 or 1-NM-PP1) and grown for 2–3 days at 25 °C or 34 °C as indicated. Colonies of cells with suppressors of pat1 alleles can be seen on YES plate at 34 °C. (b) DIC (differential interference contrast) images and DAPI staining of asci from strains described in (a).
Our pat1-as2 strain carries both hygromycin and clonat resistance markers. Therefore, these two markers cannot be easily used to introduce new alleles or constructs carrying the same drug-resistance markers. We removed the hygromycin and clonat resistance markers from the pat1-as2 strain; however, this led to decreased sensitivity to 1-NM-PP1 and poor meiotic synchrony (data not shown). We speculate that this is because DNA flanking the pat1-as2 open reading frame may affect expression levels of pat1-as2.
Materials
REAGENTS
Inhibitors (4-amino-1-tert-butyl-3-(1’-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (1-NMPP1); cat.#: A603003; 4-amino-1-tert-butyl-3-(3-bromobenzyl)pyrazolo[3,4-d]pyrimidine (3- BrB-PP1); cat. #: A602985; Toronto Research Chemicals, Inc.; http://www.trc-canada.com)
pCloneNat1 (EF101285)5, pCloneHyg1(EF101286)5 and pCloneBle1 (GQ354685)38 (these plasmids are freely available from J. Gregan's laboratory)
Anion exchange columns for purification of DNA products from PCR and for isolation of plasmid DNA
High-fidelity DNA polymerase (Pfu Turbo® DNA Polymerase; cat.#: 600250-52; Agilent Technologies, Inc.; http://www.genomics.agilent.com)
Site-directed mutagenesis kit (e.g. Agilent Technologies, Inc.; http://www.genomics.agilent.com)
Restriction enzymes (e.g. Fermentas; New England BioLabs, Inc.)
S. pombe genomic DNA (extracted from a prototrophic pat1+ strain and purified)
Dimethyl sulfoxide (Sigma)
Agarose (Sigma)
Cytox Green (Life Technologies Corp., Molecular Probes S-7020)
Hoechst 33342 (Life Technologies Corp., Molecular Probes H-3570) or DAPI (Sigma- Aldrich 32670)
Synthetic P-factor (Peptide 2.0 Inc.)
EQUIPMENT
Benchtop centrifuge (e.g., Hermle Z383K)
PCR thermal cycler
DNA gel electrophoresis unit
25 °C and 34 °C shaking incubators or water baths
flow cytometer (e.g., FACS Calibur)
fluorescence microscope
REAGENTS SETUP
For yeast cultivation, prepare standard YES medium (5 g l−1 yeast extract, 30 g l−1 glucose) supplemented with 0.15 g l−1 adenine and 0.1 g l−1 each of uracil, L-histidine, L-lysine and Lleucine. For selection, add nourseothricin (clonat) at 100 mg l−1, hygromycin B at 200 mg l−1 and zeocin at 120 mg l−1. Store the stock of drugs, in water, at −20 °C (stable for 2–3 years).
EMM2-NH4Cl medium (3.0 g l−1 potassium hydrogen phthalate, 2.2 g l−1 Na2HPO4, 1.0% (w/v) glucose, supplemented with salts, vitamins and minerals).
EMM2 medium (3.0 g l−1 potassium hydrogen phthalate, 2.2 g l−1 Na2HPO4, 5.0 g l−1 NH4Cl, 1.0% (w/v) glucose, supplemented with salts, vitamins and minerals).
YE+5S medium (5 g l−1 yeast extract, 30 g l−1 glucose, 0.15 g l−1 adenine, 0.1 g l−1 uracil, 0.1 g l−1 L-histidine, 0.1 g l−1 L-lysine and 0.1 g l−1 L-leucine).
YE+4S-Ade medium (5 g l−1 yeast extract, 30 g l−1 glucose, 0.1 g l−1 uracil, 0.1 g l−1 Lhistidine, 0.1 g l−1 L-lysine and 0.1 g l−1 L-leucine).
All media should be stored at 4 °C (stable for 1–2 years).
50× salts (52.5 g l−1 MgCl2.6H2O, 0.735 g l−1 CaCl2.2H20, 50.0 g l−1 KCl, 2.0 g l−1 Na2SO4). Store at room temperature (stable for 2–3 years).
1000× vitamins (1.0 g l−1 panthothenic acid, 10.0 g l−1 nicotinic acid, 10.0 g l−1 inositol, 10 mg l−1 biotin). Store at 4 °C in the dark (stable for 6 months).
10000× minerals (5.0 g l−1 boric acid, 4.0 g l−1 MnSO4, 4.0 g l−1 ZnSO4.7H20, 2.0 g l−1 FeCl2.6H20, 0.4 g l−1 molybdic acid, 1.0 g l−1 KI, 0.4 g l−1 CuSO4.5H20, 10.0 g l−1 citric acid). After addition of a few drops of preservative (1:1:2 chlorobenzene:dichloroethane:chlorobutane) store at room temperature (stable for 2–3 years).
Sterilize media, salts and minerals by autoclaving. Sterilize vitamins by filtration and add to media after autoclaving.
ATP analogs (e.g. 1-NM-PP1, 3-BrB-PP1) are dissolved at 10 mM in DMSO and stored at - 20 °C. Alternatively, if DMSO affects the cellular processes under investigation, dissolve ATP analogs in dimethylformamide or methanol.
Construction of the pat1-as2 strain is described in the Box 1. Genotypes of S. pombe strains are described in Table 2. All the strains and plasmids described here are freely available and can be obtained from G. Smith's or J. Gregan's labs.
Box 1. Construction of the pat1-as2 strain.
Deletion of the pat1 gene (see the Nature Protocol by Gregan et al.5 for more details)
-
1)
PCR-amplify DNA flanking the pat1 gene (SPBC19C2.05) from genomic DNA using primers 5'-AAA ATC TAG Acgc aag cgt tga ttg tcg at-3' and 5'-AAA ACT CGA Ggt ccc aat tga tgg cga aaa-3' for the upstream region and primers 5'-AAA ATC TAG Att cgt att cca aaa gct tag ttt gc-3' and 5'-AAA AAG ATC Ttc gct acc gca cgt tgt ttt-3' for the downstream region (upper case letters indicate exogenous sequences used for the cloning, and lower case letters indicate S. pombe sequences)
-
2)
Purify the products by gel electrophoresis, digest with XbaI and ligate to each other.
-
3)
Clone the heterodimer containing DNA regions flanking the pat1 gene using XhoI and BglII, into a pCloneNat1 vector (EF101285)5 carrying drug-resistance markers for E. coli (ampicillin) and S. pombe (nourseothricin).
-
4)
Amplify the resulting pCloneNat1-Δpat1 plasmid (p132) in E. coli and linearize it by cutting with XbaI
-
5)
Use the pCloneNat1-Δpat1 plasmid with selection for clonat-resistant transformants, to delete one copy of the essential pat1 gene in diploid strain JG11315.
-
6)
Confirm the deletion by colony PCR and by tetrad analysis20.
Creation of pat1-as mutants (see the Nature Protocol by Gregan et al.21 for more details)
-
7)
PCR-amplify the pat1 gene together with its promoter and terminator regions from genomic DNA using primers 5'-ATA TCT CGA Gcg att gtg ttt cct tct cat cc-3' and 5'-ATA TGG ATC Cgg tga tac aat atg act gca tgc-3'.
-
8)
Clone the amplified sequence into a pCloneHyg1 vector (EF101286)21 carrying drug resistance markers for E. coli (ampicillin) and S. pombe (hygromycin B) using XhoI and BamHI restriction sites, resulting in pCloneHyg1-pat1 plasmid (p133).
-
9)
Use site-directed mutagenesis (QuikChangeII Site Directed Mutagenesis Kit, Agilent Technologies, Inc.,) of this plasmid to change leucine 95 of Pat1, predicted to be the “gatekeeper” residue, to glycine or alanine. Oligonucleotides used for mutagenesis are as follows: 5'-aag acg cca ttt atg tcg ttg gcc agt att gtc cga atg g-3' (sense) and 5'-cca ttc gga caa tac tgg cca acg aca taa atg gcg tct t-3' (anti-sense) for the Leu95Gly mutant and 5'-aag acg cca ttt atg tcg ttg ccc agt att gtc cga atg g-3' (sense) and 5'-cca ttc gga caa tac tgg gca acg aca taa atg gcg tct t-3' (anti-sense) for the Leu95Ala mutant (mutant codons are italicized). Confirm the mutations by sequencing.
-
10)
Linearize the resulting plasmids pCloneHyg1-pat1-as(L95G) (p134) and pCloneHyg1- pat1-as(L95A) (p135) with Psp5II and transform into diploid Δpat1/pat1+ strain JG15101. Sporulate the hygromycin-resistant transformants on EMM2-NH4Cl plates at 25 °C for 36 h, and isolate haploids carrying mutant pat1-as(L95G) (further referred to as pat1-as1) or pat1- as(L95A) (further referred to as pat1-as2) alleles based on resistance to clonat and hygromycin.
-
11)
Confirm the correct integration of mutant alleles by colony PCR, and verify by sequencing the presence of the expected pat1-as alleles20.
Introducing an ectopic mat-Pc gene into pat1-as strains
-
12)
PCR-amplify lys1-mat-Pc from the pYC36 vector17 using primers 5'-ATA TTT AAT TAA ttt ttt gaa cgc taa act ttc taa g-3' and 5'-CCC CCT CGA Gaa atg att cta tcg tat cc-3'.
-
13)
Digest the amplified fragment containing lys1-mat-Pc with PacI and XhoI enzymes and clone into the pCloneBle1 vector (GQ354685)38.
-
14)
Linearize the resulting pCloneBleoMX-mat-Pc plasmid (p165) with PpuMI and integrate into the lys1+ locus of pat1-114 and pat1-as2 diploid strains by transformation using selection for bleomycin-resistant transformants. Verify the correct integration by colony PCR20.
Procedure
Preparation of the pat1-as2 strain
-
1)
Alternatively, construct the haploid pat1-as2 strain as described above and in our previous protocol21. Additional information regarding knockout construction can be found in the protocol5.
-
2)
Cross the haploid pat1-as2 strain with strains carrying ade6-210 and ade6-216 alleles to create h−pat1-as2 ade6-210 and h−pat1-as2 ade6-216 strains. Use a protoplast fusion protocol to prepare a h−/h−pat1-as2/pat1-as2 ade6-210/ade6-216 diploid strain27.
-
3)
To create diploid h− /h− cells expressing the mat-Pc gene, linearize pCloneBleoMX-mat-Pc plasmid (p165) with PpuMI restriction enzyme and transform into pat1-as2/pat1-as2 diploid strain with selection for bleomycin-resistant transformants. Confirm successful integration into the lys1 locus by PCR.
CRITICAL STEP pat1-as2 strain is temperature-sensitive. Grow cells at no more than 25 °C. Suppressors of the temperature-sensitivity arise at multiple loci11,37 ; temperature-sensitivity should be frequently checked, since the suppressed strains may not undergo meiosis.
CRITICAL STEP Diploid pat1-as2/pat1-as2 strains should be grown onYES+4S-Ade plates supplemented with 200 mg l−1 hygromycin to keep selection for diploid cells containing the pat1-as2 allele.
CRITICAL STEP Check the sensitivity of the pat1-as2 strain to 1-NM-PP1 on YE+5S medium containing 30 mM 1-NM-PP1 by comparing it to a wild-type (pat1+) strain. PAUSE POINT Strains can be stored at 4 °C for weeks or at −80 °C for years.
Arresting cells in G1 by nitrogen starvation
-
4)
Inoculate a single diploid pat1-as2/pat1-as2 colony into 5 ml of YE+4S-Ade medium and incubate overnight at 25 °C. In the morning, dilute the culture to OD600 = 0.15 (about 0.2×107 cells ml−1) in YE+4S-Ade medium and grow at 25 °C until OD600 = 0.8 – 1.0 (about 1.4- 1.8×107 cells ml−1).
-
5)
Dilute the culture into 100 ml of YE+4S-Ade medium to OD600 = 0.2 (about 0.3×107 cells ml−1) and grow at 25 °C until OD600 = 0.55 (about 0.8×107 cells ml−1).
-
6)
Centrifuge the culture (3500 rpm, 2 min, 25 °C), wash the cells twice in 100 ml of sterile water, and resuspend them in 100 ml of EMM2-NH4Cl medium.
-
7)
Incubate the culture at 25 °C for 16 – 18 h (pat1-as2/pat1-as2 (JG16022)) or for 7 h (pat1- as2/pat1-as2 mat-Pc (JG16113)) to arrest cells in G1.
CRITICAL STEP Proper G1 arrest (>80% of cells with 1n or 2n DNA content for haploids or diploids, respectively) is crucial to achieve high synchrony of meiosis.
see TROUBLESHOOTING
Inducing meiosis by inhibiting the Pat1-as2 kinase by adding 1-NM-PP1
-
8)
Centrifuge the culture (3500 rpm, 2 min, 25 °C) and resuspend the cells in 100 ml of EMM2+NH4Cl medium. (Alternatively, it may be possible to simply add NH4Cl into EMM2- NH4Cl culture instead of changing the medium. However, we have not tried this.)
-
9)
Induce meiosis by adding 1-NM-PP1 to 25 µM and incubate at 25 °C.
optional: To induce the mating pheromone response in h− /h− pat1-as2/pat1-as2 cells (JG16022), add synthetic P-factor to70 µg ml−1 to the EMM2 medium
CAUTION P-factor is expensive. It may be possible to use sxa2Δ mutation, which encodes a P factor-specific protease, to enhance sensitivity to P factor17,39.
Monitoring the progression of meiosis
-
10)
Collect 0.5 ml aliquots of culture every 60 min, centrifuge, and fix cells in 70% ethanol. PAUSE POINT fixed cells can be stored at 4 °C for several weeks.
-
11)
Stain DNA with DAPI or Hoechst 33342, examine by fluorescence microscopy, and count nuclear divisions per cell.
-
12)
Monitor pre-meiotic S phase by flow cytometry40
see TROUBLESHOOTING
Troubleshooting
- sensitivity of the pat1-as2 strain to ATP-analog is lost or decreased
-
-ATP-analog is inactive (in our hands, 1-NM-PP1 is stable in DMSO solution for several months and up to several years when stored at −20 °C)
-
-DNA flanking the pat1-as2 open reading frame have been lost or rearranged. This DNA contains several repeats which may cause instability of this region. We found that deleting drug-resistance markers flanking the pat1-as2 open reading frame led to decreased sensitivity to 1-NM-PP1 inhibitor (our unpublished data).
-
-suppressor mutations occur when pat1-as2 cells are grown at higher temperature (32–34 °C) (Figure 3a and our unpublished data). Some of these mutations may render pat1-as2 insensitive to 1-NM-PP1.
-
-
- synchrony of meiosis is low
- -
-
-strain was not a prototroph. Many auxotrophic strains fail to induce synchronous meiosis, especially those with more than one auxotrophy. Prototrophic strains should be used whenever possible, although adenine auxotrophs induce well.
-
-use prewarmed media and water bath for incubation of cell cultures to achieve higher synchrony.
-
-pat1-as2 matPc cells enter meiosis prematurely upon longer N-starvation (more than 10 hours in EMM2-NH4Cl medium).
-
-meiosis in haploid cells is abnormal (Figure 3b): unequal segregation of chromosomes as well as lagging chromosomes during anaphase are frequently observed. Therefore, errors can be easily made when scoring meiotic nuclear divisions in haploid cells.
- different results are observed in pat1-114-induced meiosis as compared to pat1-as2-induced meiosis
- -
-
-Pat1 may have roles other than its protein kinase activity18. Whereas inactivation of Pat1- as2 by adding ATP-analog is expected to inhibit the protein kinase activity only, inactivation of the temperature-sensitive Pat1-114 (or Pat1-as2) by higher temperature is likely to destroy the structure and/or stability of the Pat1 protein. This may lead to differences in meiosis induced by ATP analog-dependent Pat1-as2 inhibition and meiosis induced by shift of pat1-114 strains to higher temperature.
- different results are observed in pat1-induced meiosis when compared to normal (diploid h+/h−) meiosis
-
-although many meiotic processes occur similarly in both haploid and diploid cells, we recommend using diploid pat1-as2/pat1-as2 cells (homozygous at the mating-type locus h+/h+ or h−/h−, to prevent starvation-induced meiosis) whenever possible. This should eliminate possible artifacts caused by haploid meiosis.
-
-
Timing
The entire protocol can be completed in five days
Steps 4 – 7, Arresting cells in G1: 10 – 20 hours
Steps 8 – 9, Induction of meiosis: 8 – 14 hours
Steps 10 – 12, Monitoring meiotic progression: 1 – 2 days
Anticipated results
Using this protocol, highly synchronous meiotic cultures can be induced in the fission yeast Schizosaccharomyces pombe at optimal temperature. Using pat1-as2 mat-Pc cells offers synchronous meiosis with most tested properties similar to those of wild-type meiosis.
Acknowledgments
This work was supported by Austrian Science Fund grants P23609 and P21437 and by the Slovak Research and Development Agency under the contract No. APVV-0111-12 and APVV-0334-12. Part of this research was supported by the United States of America National Institutes of Health grant GM032194 to G.R.S. L.C. was supported by the (European Community’s) Seventh Framework Programme (FP7/2007–2013) under grant agreement number PERG07-GA-2010-268167. J.G. was supported by the (European Community’s) Seventh Framework Programme (FP7/2007–2013) under grant agreement number PCIG11-GA-2012-322300. S.P. was supported by the EMBO long-term fellowship. We thank J. Ayte, A. Yamamoto, C. Zhang, H. Murakami, A. Lorenz and Z. Benko for helpful discussions.
Footnotes
Author contribution
J.G and G.R.S. designed the experiments and wrote the manuscript. S.P, L.C. and R.H. performed experiments and contributed to writing of the manuscript. S.P, L.C. and R.H. contributed equally. Both J.G and G.R.S. are corresponding authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kerr GW, Sarkar S, Arumugam P. How to halve ploidy: lessons from budding yeast meiosis. Cell Mol Life Sci. 2012;69:3037–3051. doi: 10.1007/s00018-012-0974-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe Y. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev Mol Cell Biol. 2012;13:370–382. doi: 10.1038/nrm3349. [DOI] [PubMed] [Google Scholar]
- 4.Phadnis N, Hyppa RW, Smith GR. New and old ways to control meiotic recombination. Trends Genet. 2011;27:411–421. doi: 10.1016/j.tig.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregan J, et al. High-throughput knockout screen in fission yeast. Nat Protoc. 2006;1:2457–2464. doi: 10.1038/nprot.2006.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahler J, Schuchert P, Grimm C, Kohli J. Synchronized meiosis and recombination in fission yeast: observations with pat1-114 diploid cells. Curr Genet. 1991;19:445–451. doi: 10.1007/BF00312735. [DOI] [PubMed] [Google Scholar]
- 7.Bauer F, Matsuyama A, Yoshida M, Hermand D. Determining proteome-wide expression levels using reverse protein arrays in fission yeast. Nat Protoc. 2012;7:1830–1835. doi: 10.1038/nprot.2012.114. [DOI] [PubMed] [Google Scholar]
- 8.Ellermeier C, et al. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci U S A. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pidoux AL, Allshire RC. The role of heterochromatin in centromere function. Philos Trans R Soc Lond B Biol Sci. 2005;360:569–579. doi: 10.1098/rstb.2004.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harigaya Y, Yamamoto M. Molecular mechanisms underlying the mitosismeiosis decision. Chromosome Res. 2007;15:523–537. doi: 10.1007/s10577-007-1151-0. [DOI] [PubMed] [Google Scholar]
- 11.Nurse P. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet. 1985;198:497–502. [Google Scholar]
- 12.Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 13.Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 14.Iino Y, Yamamoto M. Negative control for the initiation of meiosis in Schizosaccharomyces pombe . Proc Natl Acad Sci U S A. 1985;82:2447–2451. doi: 10.1073/pnas.82.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;386:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. Embo J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra-Moreno A, Alves-Rodrigues I, Hidalgo E, Ayte J. Chemical genetic induction of meiosis in Schizosaccharomyces pombe. Cell Cycle. 2012;11:1621–1625. doi: 10.4161/cc.20051. [DOI] [PubMed] [Google Scholar]
- 19.Kakui Y, Sato M, Tanaka K, Yamamoto M. A novel fission yeast mei4 mutant that allows efficient synchronization of telomere dispersal and the first meiotic division. Yeast. 2011;28:467–479. doi: 10.1002/yea.1851. [DOI] [PubMed] [Google Scholar]
- 20.Cipak L, Hyppa RW, Smith GR, Gregan J. ATP analog-sensitive Pat1 protein kinase for synchronous fission yeast meiosis at physiological temperature. Cell Cycle. 2012;11:1626–1633. doi: 10.4161/cc.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregan J, et al. Construction of conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe. Nat Protoc. 2007;2:2996–3000. doi: 10.1038/nprot.2007.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tachibana-Konwalski K. Pat(ting) boosts meiosis. Cell Cycle. 2012;11:1876–1877. doi: 10.4161/cc.20513. [DOI] [PubMed] [Google Scholar]
- 23.Nosek J, Tomaska L. A new tool for an old problem: synchronizing fission yeast cells during meiosis using an ATP analog-sensitive protein kinase. Cell Cycle. 2012;11:1755–1756. doi: 10.4161/cc.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Hidalgo L, Moreno S. Chemical inactivation of Pat1: a novel approach to synchronize meiosis. Cell Cycle. 2012;11:1875. doi: 10.4161/cc.20512. [DOI] [PubMed] [Google Scholar]
- 25.Murakami H, Aiba H. Another way to induce synchronous meiosis. Cell Cycle. 2012;11:1874. doi: 10.4161/cc.20511. [DOI] [PubMed] [Google Scholar]
- 26.Wu PY. Insights from a new tool for meiotic induction in fission yeast. Cell Cycle. 2012;11:2050. doi: 10.4161/cc.20537. [DOI] [PubMed] [Google Scholar]
- 27.Sipiczki M, Ferenczy L. Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol Gen Genet. 1977;151:77–81. doi: 10.1007/BF00446915. [DOI] [PubMed] [Google Scholar]
- 28.Egel R, Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp Cell Res. 1974;88:127–1234. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- 29.Cromie GA, et al. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cromie GA, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLeod M, Stein M, Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. Embo J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto TG, Chikashige Y, Ozoe F, Kawamukai M, Hiraoka Y. Activation of the pheromone-responsive MAP kinase drives haploid cells to undergo ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J Cell Sci. 2004;117:3875–3886. doi: 10.1242/jcs.01248. [DOI] [PubMed] [Google Scholar]
- 33.Funaya C, et al. Transient structure associated with the spindle pole body directs meiotic microtubule reorganization in S. pombe. Curr Biol. 2012;22:562–574. doi: 10.1016/j.cub.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grallert A, et al. Centrosomal MPF triggers the mitotic and morphogenetic switches of fission yeast. Nat Cell Biol. 2013;15:88–95. doi: 10.1038/ncb2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay YD, Patel A, Kaemena DF, Hagan IM. Mutation of a conserved residue enhances sensitivity of analogue sensitized kinases to generate a novel approach for mitotic studies in fission yeast. J Cell Sci. 2013 doi: 10.1242/jcs.135301. Advance Online Article. [DOI] [PubMed] [Google Scholar]
- 36.Kawashima SA, Takemoto A, Nurse P, Kapoor TM. Analyzing fission yeast multidrug resistance mechanisms to develop a genetically tractable model system for chemical biology. Chem Biol. 2012;19:893–901. doi: 10.1016/j.chembiol.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iino Y, Sugimoto A, Yamamoto M. S. pombe pac1+, whose overexpression inhibits sexual development, encodes a ribonuclease III-like RNase. Embo J. 1991;10:221–226. doi: 10.1002/j.1460-2075.1991.tb07939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spirek M, et al. S. pombe genome deletion project: An update. Cell Cycle. 2010;9:2399–2402. doi: 10.4161/cc.9.12.11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 1994;8:328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- 40.Sabatinos SA, Forsburg SL. Measuring DNA content by flow cytometry in fission yeast. Methods Mol Biol. 2009;521:449–461. doi: 10.1007/978-1-60327-815-7_25. [DOI] [PubMed] [Google Scholar]
- 41.Shimanuki M, et al. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells. 2007;12:677–692. doi: 10.1111/j.1365-2443.2007.01079.x. [DOI] [PubMed] [Google Scholar]
- 42.Takeda K, et al. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc Natl Acad Sci U S A. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu BC. Dark dependence of meiosis at elevated temperatures in the basidiomycete Coprinus lagopus. J Bacteriol. 1972;111:833–834. doi: 10.1128/jb.111.3.833-834.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malik CP. Effect of variations in temperature on meiosis in Gagea reticulata schultes. Nature. 1960;187:805–806. doi: 10.1038/187805b0. [DOI] [PubMed] [Google Scholar]
- 45.Francis KE, et al. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:3913–3918. doi: 10.1073/pnas.0608936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loidl J. Effects of elevated temperature on meiotic chromosome synapsis in Allium ursinum. Chromosoma. 1989;97:449–458. [Google Scholar]