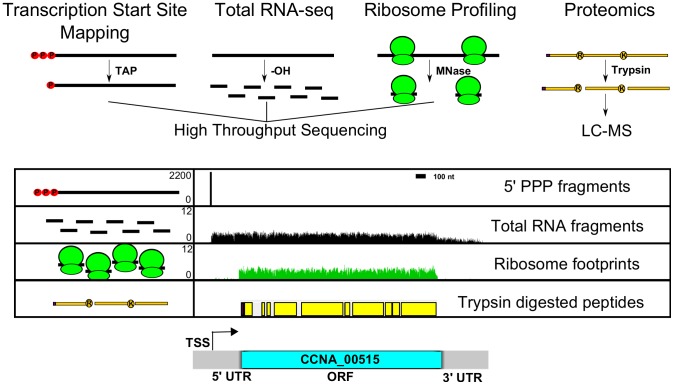
Figure 1. Genome-wide data set integration to map the genetic elements in the C. crescentus genome.
5′PPP transcription start site (TSS) (Zhou et al. [unpublished data]) (red spheres with black bar), RNA-seq density (black bars), ribosome footprints (green ribosomes), and LC-MS peptide coverage [7] (yellow bars) shown for a single gene (CCNA_00515) between 528700 and 532200 bp. 5′PPP data generated from Tobacco Acid Pyrophosphate (TAP) enriched 5′ global RACE. 5′ PPP fragments plotted with Y-axis scale in #reads. Base hydrolyzed (-OH) RNA-seq data plotted with Y-axis scale in log(#reads+1). Micrococcal nuclease (MNase) protected ribosome footprints plotted with Y-axis scale in log(#reads+1). LC-MS-identified tryptic peptides are mapped onto their respective positions of the CDS with potential ribosomal initiated N-terminal residue in purple. Respective genomic features are highlighted including transcriptional start site (TSS), 5′ untranslated region (UTR), coding region (blue bar), and 3′ UTR of the expressed element. Y-axis scales are similar in all subsequent figures.