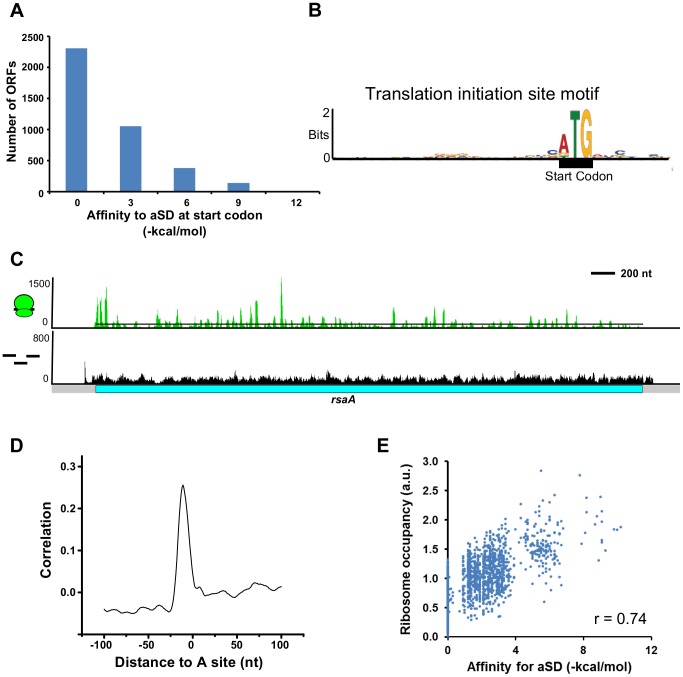
Figure 3. Role of the Shine-Dalgarno sequence in translation initiation and pausing.
A. Global lack of SD sites in front of start codons. mRNA affinity to the aSD site on the ribosome was calculated using the Free2bind package [16]. −4.4 kcal/mol is the cutoff for SD identification based on the predicted annealing between the aSD and translation initiation region as in [13]. B. Translation initiation site motif derived from all start codons in the genome generated in MEME [77]. C. Ribosome occupancy profiles reveal pausing identified by peaks of ribosomes above the average read density (black line). Stronger pauses are shown by a larger peak height. Y-axis value is #reads on linear scale. D. Plot of the normalized cross-correlation function between pauses in the ribosome occupancy profiles and the presence of SD sequences. The plot is centered at the A-site of ribosome pauses and the peak of correlation occurs in proximity to the aSD site on the ribosome. E. Plot of SD binding affinity for the aSD compared to the occupancy of ribosomes translating them. r is the correlation coefficient.