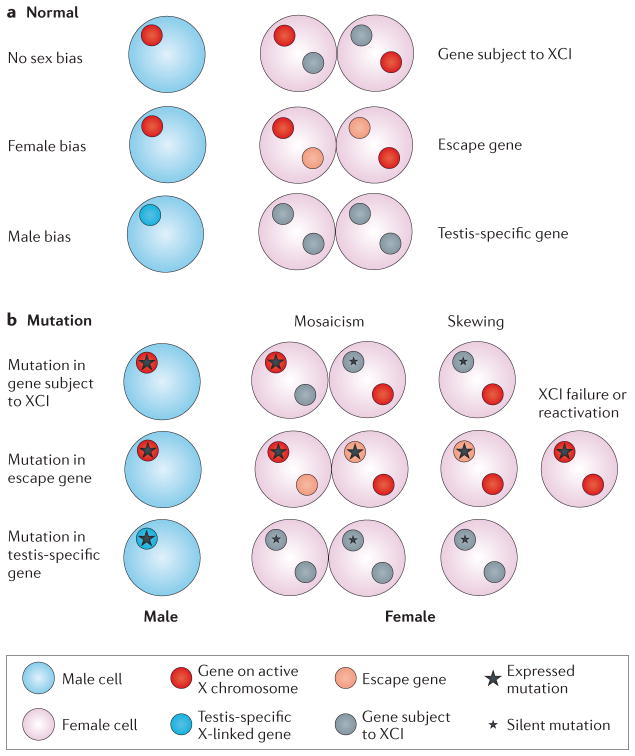
Figure 4. Variability of X-linked gene expression and sex bias.
a | Sex bias in X-linked gene expression in normal individuals is shown. There is no sex bias for genes that are subject to X chromosome inactivation (XCI) or for genes with equivalent X–Y paralogues (not shown). A female bias occurs for genes that escape XCI but that have no equivalent Y paralogue; a male bias occurs for genes that are solely expressed in male organs such as the testes. b | The effects of X-linked gene mutation depend on XCI patterns. For genes that are subject to XCI, a mutation that affects males does not necessarily affect females, who can be rescued either by random XCI (which leads to mosaicism for cells with the mutant allele silenced) or by selective skewing in favour of cells that express the normal allele. For escape genes, males can be partly rescued by expression of a Y paralogue with similar function (if it exists), whereas females will be haploinsufficient even if there is skewing in favour of the normal allele. Severity of the haploinsufficiency in females depends on the expression levels from the inactive X chromosome. Mutations in genes that are essential for XCI (such as the gene that encodes X inactive specific transcript (XIST)) specifically affect females by causing XCI failure. If a mutation is in an X-linked gene that is solely expressed in male organs (such as the testes), only males will be affected. However, as many of the testis-specific genes are ampliconic, the effect of a mutation in one copy will be partly rescued by the normal copies.