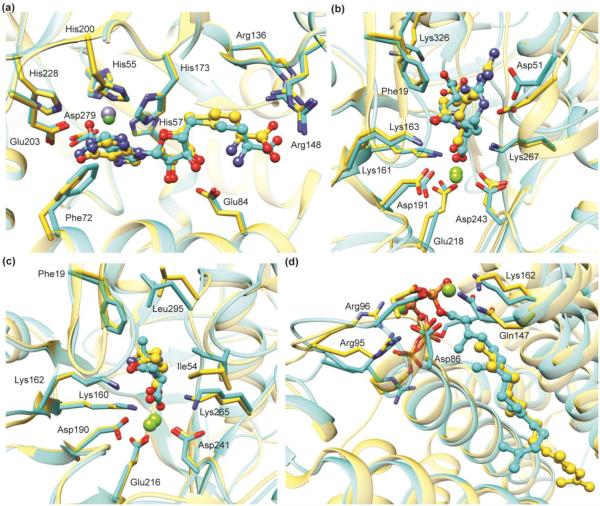
Figure 2.
Predicted binding poses are in good agreement with subsequently determined experimental structures. Predicted ligand binding mode (cyan) superimposed with the X-ray crystal structure (gold) of: (a) S-adenosylhomocysteine deaminase (PDB: 2PLM); (b) N-succinyl-L-Arg racemase (PDB: 2P8C); (c) D-Ala-D-Ala epimerase (PDB: 3Q4D), and (d) a polyprenyl synthase (PDB: 4FP4). In (b), (c), and (d), the docking predictions were made using homology models based on crystal structures with 35%, 39%, and 29% sequence identity, respectively.