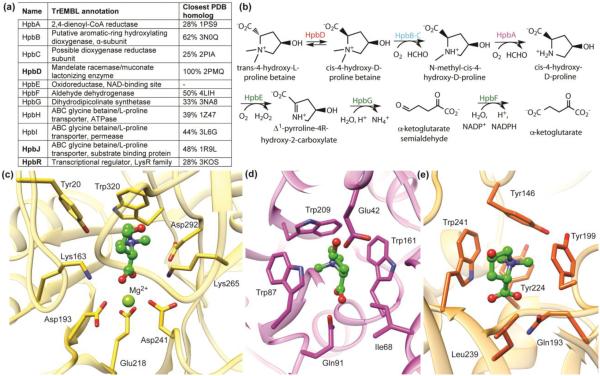
Figure 3.
Structure-guided discovery of new enzymes in a novel hydroxyproline betaine metabolism pathway. Panel (a) shows the name, TrEMBL annotation, and most similar homolog in the PDB for each protein in the pathway. The automated TrEMBL annotations are incorrect or imprecise for all proteins in the pathway. However, there is rich structural information that can be used for modeling and docking, as shown in the closest PDB homolog column. The pathway is shown in (b). Panels c-e show the binding site and/or active site of the three proteins (HpbD, HpbJ and HpbR, shown in bold in (a)) in the pathway, respectively, along with the docking-predicted binding mode for the ligand trans-4-hydroxy-L-proline betaine (ball-and-stick, green color). Both HpbJ and HpbR have a predicted cation-π cage, known for binding quaternary amines. In HpbD, two catalytic residues (Lys163 and Lys265) replace aromatic residues, leaving Trp320 as the key aromatic residue forming a cation-π interaction with the substrate.