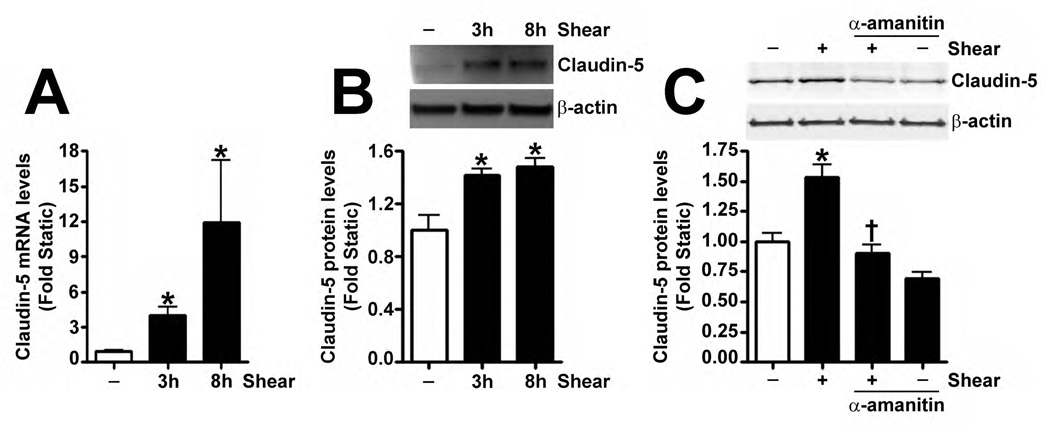
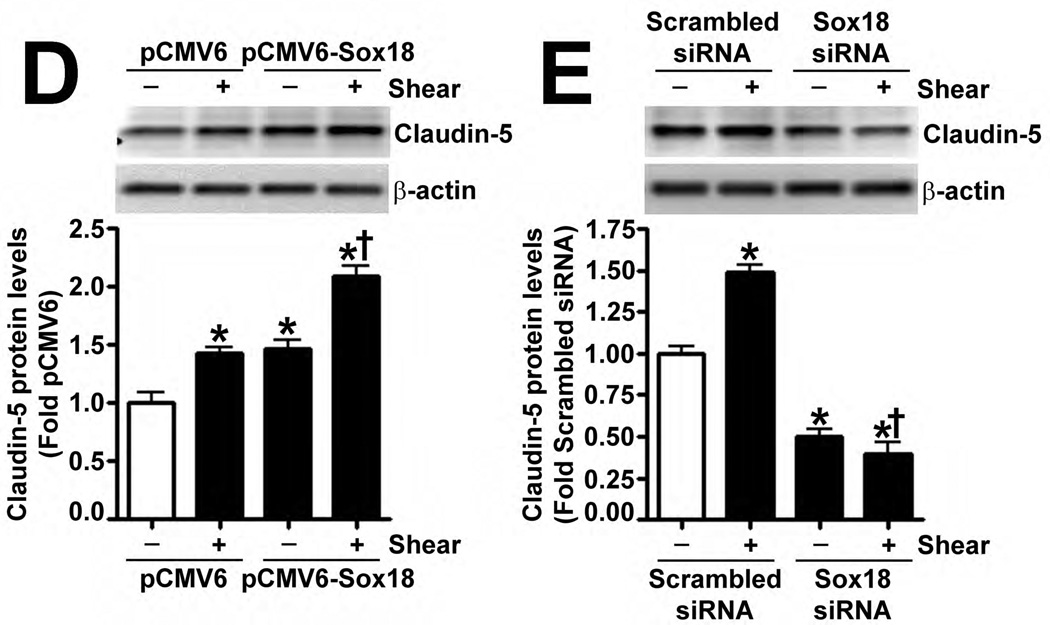
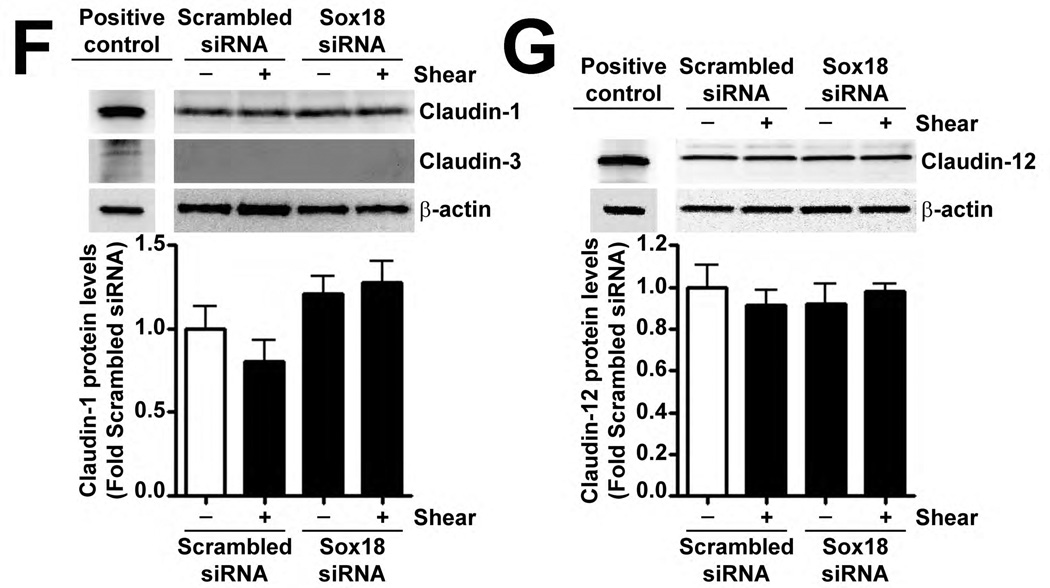
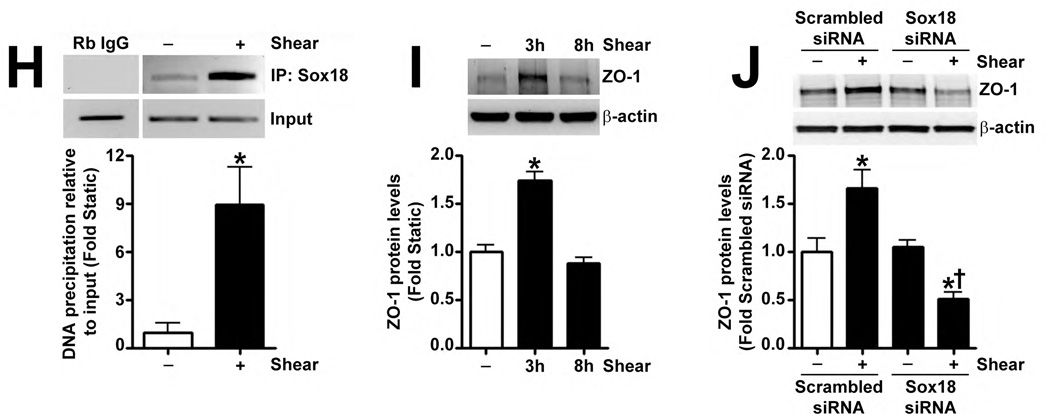
Fig. 4. Shear stress up-regulates the cellular tight junction proteins, Claudin-5 and Zonula occludens-1, in a Sox18 dependent manner in pulmonary artery endothelial cells.
PAEC were subjected to static conditions or 20 dyn/cm2 of shear stress for 3 and 8 h. Messenger RNA levels for Claudin-5 were significantly higher in PAEC exposed to shear compared to static culture conditions, as determined by SYBR Green real-time RT-PCR analysis (A). Similarly, immunoblot analysis indicated that Claudin-5 protein levels were significantly higher in PAEC exposed to 3h and 8h of 20 dyn/cm2 of shear stress (B). The increase in Claudin-5 protein levels after 3 h of shear was abrogated by 24 h pre-treatment with the transcriptional inhibitor, α-amanitin (C). PAEC transfected with pCMV6-Sox18 or Sox18 siRNA for 48 h were exposed to 3 h of 20 dyn/cm2 of shear stress. Immunoblot analysis showed that the over-expression of Sox18 potentiates (D), while the knockdown of Sox18 attenuates (E), the shear mediated increase in Claudin-5 levels. Claudin-1 (F) and Claudin-12 (G) protein levels were not altered by Sox18 depletion in the presence or absence of shear. Claudin-3 was not detected (F). Chromatin immunoprecipitation indicated that shear stress induces a significant increase in the binding of Sox18 to the endogenous Claudin-5 promoter (H). Immunoblot analysis also indicated that Zonula occludens-1 (ZO-1) protein levels were significantly increased in PAEC exposed to 3h of 20 dyn/cm2 of shear stress compared to static controls (I). In addition, PAEC were transfected with Sox18 siRNA for 48 h and exposed to 3 h of 20 dyn/cm2 of shear stress. Immunoblot analysis showed that the silencing of Sox18 attenuates the shear mediated increase in ZO-1 protein levels (J). Values are mean ± SEM, n=4–6. *p<0.05 vs. Static (A-C, H, I), pCMV6 (D), Scrambled siRNA (E-G, J); † P<0.05 vs. pCMV6+Shear (D) or Scrambled siRNA+Shear (E, J).