Figure 1.
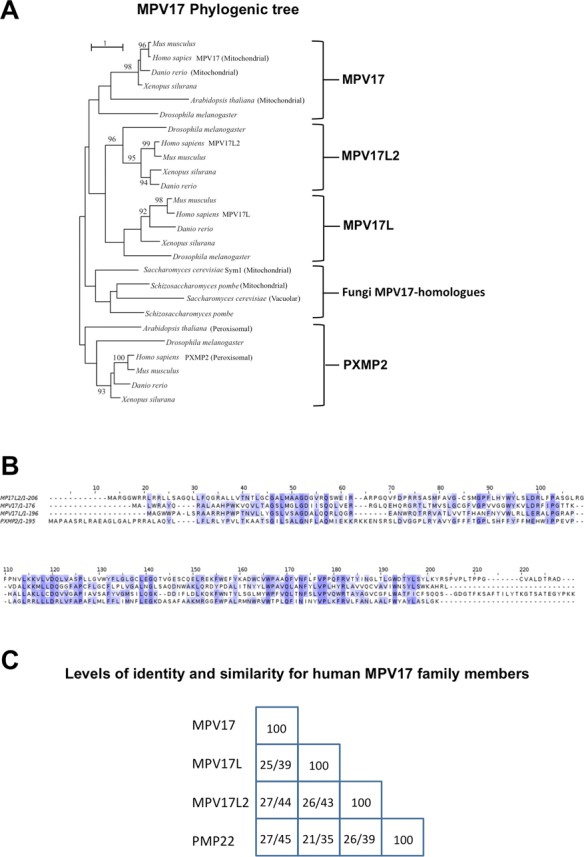
The MPV17 family of proteins. (A) Maximum likelihood phylogeny of the MPV17 protein family, with an emphasis on the origin of the human members of the protein family. Experimental localization data for proteins are indicated with the branches. Bootstrap values above 90 out of 100 are shown. The phylogeny indicates an early separation of the MPV17 clade from the PXMP2 clade, and a later, metazoan origin of the MPV17L and MPV17L2 clades. Overall there is a general pattern of lineage specific duplications throughout the eukaryotes, leading, for example, to 10 members of the MPV17 protein family in Drosophila melanogaster (not all of which are shown). Protein location within an orthologous group tends to be conserved, but gene duplications often leads to the acquiring of a new location (60), as can be observed for the PXMP2/MPV17 gene duplication at the origin of the eukaryotes and the gene duplication within the fungi that potentially give rise to a new, vacuolar location of YOR292C (61). (B) Sequence alignment of the four members of the human MPV17 family. Alignment was based on a ClustalX alignment of the protein family. Colour coding of the level of conservation was based on Jalview (62). (C) Levels of sequence identity/similarity among the human MPV17 family members, the ordering of the proteins along the horizontal axis is identical as that along the vertical axis. Pairwise levels of sequence identity/similarity were based on global pairwise sequence alignment, using default parameters in the EMBOSS package (63). MPV17L2 has a slightly higher level of sequence identity and sequence similarity to MPV17 than MPV17L has to MPV17.
