Figure 5.
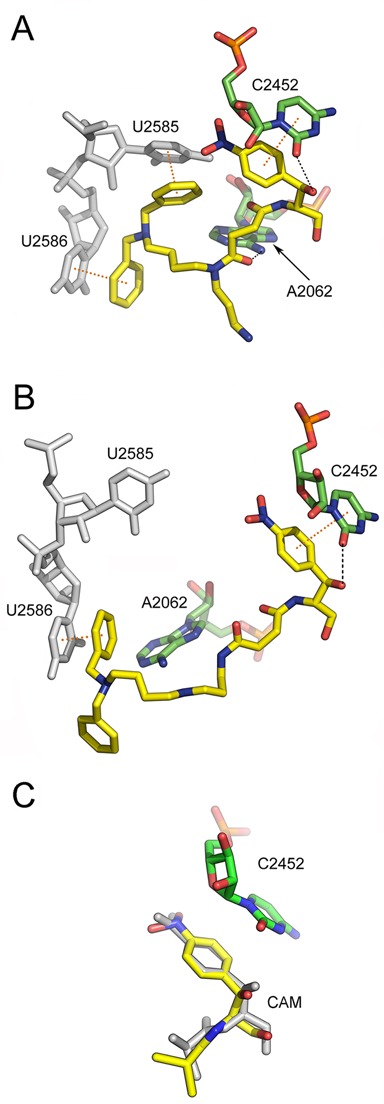
Binding position of two PA–CAM conjugates on the Escherichia coli ribosome, as detected by Molecular Dynamics Simulations. The ligand models have been docked into the 50S ribosomal subunit, by positioning their CAM moiety within the CAM crystallographic pocket (31). (A) Binding position of compound 4 (yellow); hydrogen bonding with A2062 and C2452 is shown by black dashes, while π-stacking arrangements with U2585 and U2586 (gray) are indicated by yellow dots connecting the centers of the interacting aromatic rings. Other residues of 23S rRNA placed adjacently to the binding pocket of 4 are ignored for clarity. (B) Binding position of compound 5 (yellow); a π-stacking interaction of 5 with U2586 (gray) is shown at the left. (C) Overlay of CAM structures from MD simulation (yellow) and crystallographic analysis (gray; PDB:3OFC). Other residues of 23S rRNA placed adjacently to the binding pocket of CAM, except for C2452, are ignored for clarity.
