Figure 1.
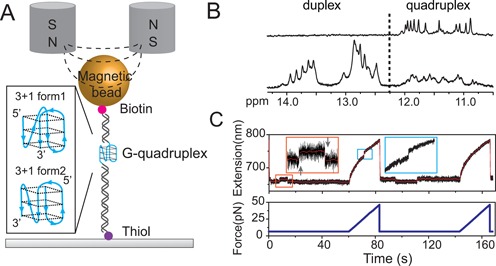
Experimental setup and procedure. (A) Schematic diagram of G4 DNA and magnetic tweezers. The 26 mer ssDNA of four-repeat human telomeric sequence d(TTGGG(TTAGGG)3TTT) is sandwiched bewteen a lower 1449 bp and an upper 601 bp dsDNA handles. The DNA construct is tethered between a 2.8 μm-diameter paramagnetic bead via biotin-streptavidin linkage and an amine functionalized coverslip surface through covalent cross-linker sulfo-SMCC. Inset shows two possible hybrid-type G4 structures that may form on the sequence (16,17). (B) Imino proton NMR spectrum of telomeric G4 sequence d(TTGGG(TTAGGG)3TTT) (top) and the 1:1:1 mixture of three sequences, d(CGAGTCTGTGCACAAGGTGC), d(CTACTGACCTGGCTGC) and d(CTTGTGCACAGACTCGTTGGG(TTAGGG)3TTTGCAGCCAGGTCAGTAGCGAC) (bottom). The mixture is expected to form a G-quadruplex in the centre flanked by two 16-bp Watson–Crick duplexes at the 5′- and 3′-ends respectively (underlined sequences). (C) Typical force responses of G4 DNA in two repeating stretching cycles (original and smoothed extension data are shown in black and red, respectively). In each cycle, a constant force was maintained at 6.5 pN for 60 s then increased to 50 pN at a constant loading rate of 2 pN/s. In the first cycle, at the constant force of 6.5 pN, two extension states with an extension difference of ∼6 nm were observed, indicated by an unfolding transition (up-arrow) followed by a refolding transition (down-arrow) in the zoom-in inset in the orange rectangle. During the subsequent force-increase scan at 2 pN/s, a typical G4 unfolding indicated by a sudden extension jump with a step size of ∼8 nm occurred at ∼25 pN (marked in cyan rectangle). In the second cycle after force was jumped back to 6.5 pN, a refolding transition and a following unfolding transition were observed. In the subsequent force-increase scan at 2 pN/s, G4 unfolding was not observed.
