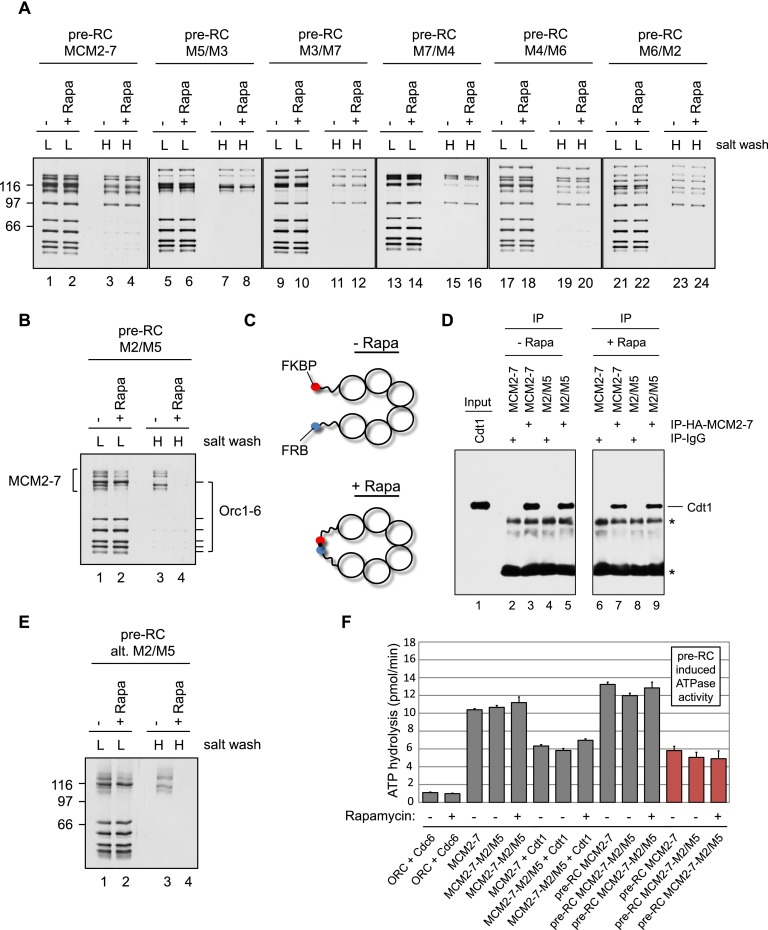
Figure 3.
Only MCM2–7–M2/M5 in the presence of rapamycin displays a helicase loading defect. (A) Pre-RC reactions using MCM2–7 and five MCM2–7–FKBP/FRB protein complexes were assembled in the presence or absence of rapamycin and ATP, washed with low-salt (L) or high-salt (H) buffer, and analyzed by silver staining. The smallest subunit of the Orc1–6 complex stains only weakly by silver staining. (B) Pre-RC reactions using MCM2–7–M2/M5 were assembled in the presence or absence of rapamycin and ATP, washed with low salt (L) or high salt (H), and analyzed by silver staining. (C) Illustration showing the rapamycin-induced linkage between neighboring Mcm subunits. (D) The MCM2–7–Cdt1 interaction analysis used IgG control beads or MCM2–7 (HA-Mcm3) coupled to anti-HA beads. A 30% input is shown along with 100% of the immunoprecipitate. Asterisks mark nonspecific IgG-related bands. (E) Pre-RC reactions using an alternative MCM2–7–M2/M5 construct were assembled in the presence or absence of rapamycin and ATP, washed with low salt (L) or high salt (H), and analyzed by silver staining. (F) Analysis of ATPase activities during pre-RC assembly with wild-type (wt) MCM2–7 or MCM2–7–M2/M5 in the presence or absence of rapamycin. The ATP hydrolysis rates were determined for the indicated proteins in the presence of ARS1 origin DNA. Together, ORC, Cdc6, Cdt1, and MCM2–7 induced a strong ATPase activity. The pre-RC-induced ATPase activity is shown in red (ATPase activity of the full pre-RC reaction with the ATPase activity of the individual components [ORC/Cdc6 + Cdt1/MCM2–7] subtracted).