Abstract
Background
All modern rosids originated from a common hexapolyploid ancestor, and the genomes of some rosids have undergone one or more cycles of paleopolyploidy. After the duplication of the ancient genome, wholesale gene loss and gene subfunctionalization has occurred. Using the extensin super-gene family as an example, we tracked the differential retention and expansion of ancestral extensin genes in four modern rosids, Arabidopsis, Populus, Vitis and Carica, using several analytical methods.
Results
The majority of extensin genes in each of the modern rosids were found to originate from different ancestral genes. In Arabidopsis and Populus, almost half of the extensins were paralogous duplicates within the genome of each species. By contrast, no paralogous extensins were detected in Vitis and Carica, which have only undergone the common γ-triplication event. It was noteworthy that a group of extensins containing the IPR006706 domain had actively duplicated in Arabidopsis, giving rise to a neo-extensin around every 3 million years. However, such extensins were absent from, or rare in, the other three rosids. A detailed examination revealed that this group of extensins had proliferated significantly in the genomes of a number of species in the Brassicaceae. We propose that this group of extensins might play important roles in the biology and in the evolution of the Brassicaceae. Our analyses also revealed that nearly all of the paralogous and orthologous extensin-pairs have been under strong purifying selection, leading to the strong conservation of the function of extensins duplicated from the same ancestral gene.
Conclusions
Our analyses show that extensins originating from a common ancestor have been differentially retained and expanded among four modern rosids. Our findings suggest that, if Arabidopsis is used as the model plant, we can only learn a limited amount about the functions of a particular gene family. These results also provide an example of how it is essential to learn the origination of a gene when analyzing its function across different plant species.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-612) contains supplementary material, which is available to authorized users.
Keywords: Extensin, Paleopolyploid, Purifying selection, Syntenic chromosomal block, Gene duplication manners
Background
During the evolutionary process, whole-genome duplications (WGDs) have recurred in many lineages of the angiosperms [1], leading to remarkable fluctuations in their genome sizes. Following WGDs, wholesale gene loss [1] and gene subfunctionalization [2, 3] can occur. Synteny and collinearity analyses of plant genomes have suggested that an ancient genome triplication (γ-triplication) event occurred in the common ancestor of Vitis, Arabidopsis, Carica, and Populus, resulting in a paleohexaploid [4]. After the γ-triplication event, Arabidopsis was affected by two recent paleopolyploidy events: β- and α- duplications. The latter was the most recent, occurring approximately 40 million years (MYs) ago [5]. In Populus, there was a duplication event specific to its own salicoid lineage (P-duplication), which occurred between the β- and α- duplication events about 65 MYs ago [6]. By contrast, in Vitis (grape) and Carica (papaya), there was only the common γ-triplication event and no subsequent WGDs [4, 7]. Ancient polyploidization events have affected the number of genes in various gene families. Tracking the differential retention and expansion of ancestral genes in modern plants is critical to learn the function of homologous genes across different plant species. In this study, we demonstrate the differential retention and expansion of ancestral genes in four rosids, focusing on the extensin super-gene family as an example.
Extensins are hydroxyproline-rich glycoproteins (HRGPs), and are members of a superfamily of plant cell-wall proteins that includes arabinogalactan proteins, extensins, and proline-rich proteins [8]. Extensins account for 1–15% of the dry weight of the cell wall of dicots [9]. In terms of their amino acid compositions, extensins are rich in hydroxyproline (Hyp), serine (Ser), and contain various amounts of tyrosine, valine, lysine, and histidine. Extensin, in a narrow sense, describes HRGPs with the characteristic Ser-Hyp-Hyp-Hyp-Hyp motif [10]. Recently, however, Showalter et al. [8] identified putative extensins with two or more repeats of the standard Ser-Pro-Pro-Pro and/or Ser-Pro-Pro-Pro-Pro sequences.
Extensins are fully functional after post-translational modification. The proline residues of extensins are first hydroxylated to Hyp, and then modified by arabinosyltransferases [11]. After being secreted into the cell wall, mature extensins form a network by oxidative cross-linking of several Tyr residues [12, 13]. Extensins have been implicated in nearly all aspects of plant growth and development [14]. Studies have shown that pollen- and pistil-specific extensin-like proteins play roles in reproduction [15–17]. Extensins are also involved in the responses to wounding and pathogen invasion [18, 19]. In recent years, studies on the Arabidopsis rsh mutant have demonstrated that extensins not only play an essential role in strengthening mature cell walls, but also in shaping the cell, positioning the cell plate during cytokinesis, and allowing normal embryo development [20]. The RSH extensin (AtEXT3) was thought to function exclusively in the cessation of cell growth, but recent research showed that it also has an essential role in the initiation of new cell growth [20].
Extensin genes are a special research interest in plant biology because of their biological importance. In this study, focusing on the extensin super-gene family as an example, we analyzed the expansion of these genes in Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera, and Carica papaya, whose genomes have been affected by one or more paleopolyploidy events. We aimed to track the differential retention and expansion of the ancestral extensins in these four morden rosids to provide a panoramic view of the evolutionary process of a super-gene family.
Methods
Identification of extensins
Extensins were identified following the method described by Showalter et al. [8]. First, we searched for two or more SP3/SP4 repeats in protein sequences in genomic databases of Arabidopsis thaliana (TAIR10 release of November 2010; http://www.arabidopsis.org/), Populus trichocarpa (JGIv3.0, ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Ptrichocarpa/), Vitis vinifera (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Vvinifera), and Carica papaya (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Cpapaya/). The protein hits were subsequently scanned by InterPro (European Bioinformatics Institute) [21, 22] to find signature protein domains, including IPR006706 (extensin-2), IPR006041 (pollen Oie e 1 allergen/extensin), IPR003882 (pistil-specific extensin-like protein), IPR003883 (extensin-1), PR01217 (proline-rich extensin), and PTHR23201 (extensin, proline-rich protein).
Identification of paralogs and orthologs
Paralogs and orthologs were identified following the method described by Blanc and Wolfe [3]. For each species, all-against-all nucleotide sequence similarity searches were performed among the transcribed sequences using BLASTN software [23]. Sequences that aligned over 300 bp and showed at least 40% identity were defined as pairs of paralogs. To identify putative orthologs between two species (A and B), each sequence from species A was searched against all sequences from species B using BLASTN. Additionally, each sequence from species B was searched against all sequences from species A. The two sequences were defined as orthologs if each of them was the best hit of the other, and if more than 300 bp of the two sequences aligned.
Calculation of ω and Ks values
Pairwise protein sequence alignment was performed using MAFFT v6.8 [24, 25]. Then, the protein alignments were re-edited into codon-based alignments using an in-house PERL script. The codon-based alignments were converted into TREE format files using ClustalX [26] and a PAML-compatible format using DAMBE [27]. The PAML [28] -format files were further converted into NUC format. A ‘bin’ folder was created, and the data files (TREE-format file and NUC-format files) and PAML executive programs (codeml.exe, codeml.ctl) were copied into the ‘bin’. Finally, codeml.exe was run to generate the ω, dN, and dS values, where ω = dN/dS and dS = Ks.
Phylogenetic trees construction
Protein sequences of the extensins in the four plant species were aligned using the L-INS-i software implemented in MAFFT v. 6.8 [24, 25] with the following parameters: the scoring matrix for amino acid sequences was BLOSUM62, the gap opening penalty was 2.0, and the gap extension penalty was 0.2. The derived protein alignments were re-edited into codon-based alignments using an in-house PERL script. Phylogenetic trees were reconstructed with MEGA v. 5.0 [29] using the minimum evolution (ME) and neighbour-joining (NJ) methods. The reliability of interior branches was assessed with 1,000 bootstrap re-samplings.
We constructed other phylogenetic trees using more advanced methods, including the maximum likelihood (ML) and Bayesian inference (BI) methods. The ML tree was generated with RAxML using the GTR+G model and nucleotide data sets [30]. The BI tree was generated with PhyloBayes-MPI [31] using the GTR-CAT+G4 model and nucleotide data sets. For each data set, two independent runs were executed until the maximum discrepancy between the bi-partition was less than 0.1. In both the ML and BI trees, the reliability of interior branches was assessed with 1,000 bootstrap re-samplings. On the established trees, branches supported with bootstrap values greater than 60% were joined.
Analysis of duplication manners for extensin genes
The plant genome duplication database (PGDD; available at http://chibba.agtec.uga.edu/duplication/) is a public database to identify and catalogue plant genes in terms of intra-genome or cross-genome syntenic relationships. To identify extensin genes that had arisen from segmental or whole-genome duplications (S/WGD) in the genomes of the four rosid plants, we used the PGDD [32] to retrieve the syntenic chromosomal blocks (SCBs) associated with the expansion of extensin genes through S/WGDs. First, in the PGDD, we detected all of the chromosomal blocks that contained extensin genes in each of the four rosids. Then, we retrieved their syntenic blocks within and between species from the PGDD. The extensin genes duplicated through S/WGDs were identified based on gene collinearity on SCBs. In this analysis, the counterparts of a particular extensin gene on an SCB may have been retained as extensins, subfunctionalized into non-extensins, or completely lost. Additionally, some of the syntenic blocks could have been completely lost after the ancient S/WGDs.
Extensin genes expanded through tandem duplication (TD) were inferred following the method described by Tuskan et al. [6], with a Smith-Waterman alignment E value of ≤ 10-25 and a 100-kb window. If the paralogous extensins had expanded neither through W/SGD nor TD, they were considered to have proliferated via other duplication strategies [33].
Reconstructing a tentative phylogeny of the large paralogous group in Arabidopsis
We used a hierarchical clustering method to reconstruct a tentative phylogeny of the large paralogous group in Arabidopsis, as follows [3]: (1) all sequences in the group were treated as separate clusters; (2) the Ks values for all possible pairs of clusters were compared; (3) the pair of clusters with the smallest Ks value was replaced by a single new cluster containing all of their sequences; (4) the median Ks value was chosen to represent the duplication event that gave rise to the two merged clusters; and (5) steps 2–4 were repeated until all sequences were contained in a single cluster. When two clusters (A and B) contained more than one sequence, their associated Ks value in step 2 was the median Ks obtained for all possible pairs of any sequence from A and B. The gene neutral evolutionary rate in Arabidopsis was reported to be 1.5 × 10-8 [34]. The duplication times for the paralogous extensin pairs were estimated by Ks/(2 × 1.5 × 10-8) according to Blanc and Wolfe’s calculation [3].
Results
Extensingenes in four modern rosids
The four studied rosids, A. thaliana, P. trichocarpa, C. papaya, and V. vinifera, originate from a common paleohexapolyploid ancestor. Based on the WGDs in each species, the multiplicity ratio for an ancestral gene in the above species should be 4:2:1:1. We retrieved 46, 37, 18, and five putative extensin genes from the genomes of A. thaliana, P. trichocarpa, C. papaya, and V. vinifera, respectively (Additional file 1). In V. vinifera, 94.5% of its genome demonstrates the occurrence of the paleohexaploidy, and this species appears to have the closest karyotype to that of the common ancestor [7]. When V. vinifera was taken as the baseline, the current ratio of extensin genes among the four genes was 9.2: 7.4: 3.6: 1. Compared with V. vinifera, the other three species showed higher-than-expected current ratios of extensins. Therefore, V. vinifera may have lost extensin genes faster than did the other species, or extensins may have expanded faster in the other three species than in V. vinifera.
A detailed examination (Additional file 2) revealed that the most common type of extensins were those containing the PR01217 (PRICHEXTENSN) domain. This group made up the largest proportion of extensins in each of the four modern rosids, accounting for 54%, 89%, 95%, and 80% of all of the extensins in Arabidopsis, Populus, Carica and Vitis, respectively. Arabidopsis has 18 extensins containing the IPR006706 (Extensin-2) domain; this group has specifically and remarkably proliferated only in Arabidopsis (accounting for 39.1% of all extensins in Arabidopsis). This group of extensins was rare or absent from the other three rosids.
Extensinparalogs and orthologs
The paralogous and orthologous extensins within and between species are listed in Table 1. In Arabidopsis, five paralogous groups containing 22 extensins were identified. In Populus, there were seven paralogous groups containing 18 extensins. Thus, nearly half of the extensins were associated with intra-specific duplication in Arabidopsis and Populus. By contrast, there were no paralogous duplicates in Carica and Vitis, which have undergone only the common γ triplication event and no subsequent WGDs. There was a large paralogous group containing 12 extensins in Arabidopsis (Table 1), all encoding proteins with the IPR006706 (Extensin-2) domain.
Table 1.
Paralogous and orthologous extensins identified in four modern rosid plants
| Arabidopsis thaliana | Populus trichocarpa | Carica papaya | Vitis vinifera | |
|---|---|---|---|---|
| Paralogous extensins within species | A2-A6-A7-A8-A9-A10-A11-A12-A13-A15-A16-A17; A18-A19-A20; A24-A28-A49; A25-A31; A26-A27 | P1-P2; P5-P7-P6; P10-P12; P18- P19; P20-P22-P28; P26-P17; P34-P35-P36-P37 | — | — |
| Orthologous extensins between species | P5-A35; P6-A39; P29-A44; C18-P30; C6-P6; C7-P24; C8-P20; V1-C6; V1-P5 | |||
The observed ratio of extensins (37:18) was close to the expected ratio (2:1) based on the paleopolyploidy of Populus and Carica. These two species shared the most orthologous pairs (Table 1), followed by Populus and Arabidopsis. Although Carica was the closest relative of Arabidopsis among the four species (both belong to the Brassicales), they had no orthologous extensins. There were also no orthologous extensins between Vitis and Arabidopsis. One pair of orthologous extensins was detected between Carica and Vitis.
The ratio of non-synonymous substitutions per non-synonymous site (dN) to the synonymous substitutions per synonymous site (dS) is an indicator of natural selection [35]. We can evaluate the driving force shaping the evolution of extensins based on the value of ω = dN/dS. If ω < 1, the gene is undergoing purifying selection, resulting in gene function becoming more convergent. When ω > 1, gene function will be more divergent. Our calculations (Additional file 3) clearly showed that all the paralogous and orthologous extensins within and between species were under strong purifying selection, with an overall mean value of ω = 0.15. Purifying selection was not evident for only one paralogous pair, P22-P28, in Populus (ω = 0.9765). The average ω for paralogous pairs was 0.24 in poplar, and 0.12 in Arabidopsis. Therefore, paralogous pairs of extensins have been under stronger purifying selection in Arabidopsis than in poplar.
Phylogenetic analysis of extensingenes in four modern rosids
Phylogenetic trees were constructed with MEGA [29] using the ME method (Figure 1) and the NJ method (Additional file 4). The ME and NJ trees showed identical topologies. A total of 106 genes were distributed among 43 branches with bootstrap values ≥60%, and formed three distinct clades (Figure 1). Clade I consisted of 16 extensins from Arabidopsis. This clade consisted of two sub-clades, one containing A20, A22, A18 and A19, and the other containing the large paralogous group of 12 Arabidopsis extensins described above. The fact that all of the extensins in this clade were from Arabidopsis, combined with the results of the paralog analysis (Table 1), indicated that all of the extensins in clade I were intra-specific duplicates in Arabidopsis. Clade II consisted of 12 extensins; two from Arabidopsis, six from Populus, three from Carica, and one from Vitis. Extensins in this clade included two paralogous groups from Populus (P1-P2 and P5-P6-P7) and six orthologous pairs shared by the four modern rosids (C7-P24, P5-A35, P6-A39, C6-P6, V1-P5 and V1-C6). Therefore, most of the extensins in this clade represented the ancestral relics shared between species. The extensins in this clade had expanded most dramatically in Populus. Clade III contained 22 extensins: nine from Arabidopsis, 10 from Populus, two from Carica, and one from Vitis. Paralogous groups in this clade were A24-A28-A49, A26-A27, and A25-A31 from Arabidopsis, and P20-P22-P28, P18-P19, P17-P26, and P10-P12 from Populus. Clade III contained only one orthologous pair, C8-P20. Therefore, most of the extensins in this clade were intra-specific duplicates that have expanded in Arabidopsis and Populus. As well as the three distinct clades described above, there was a small clade containing four paralogous extensins from Populus (P34, P35, P36, and P37). These represented extensins that have specifically expanded within the Populus genome.
Figure 1.
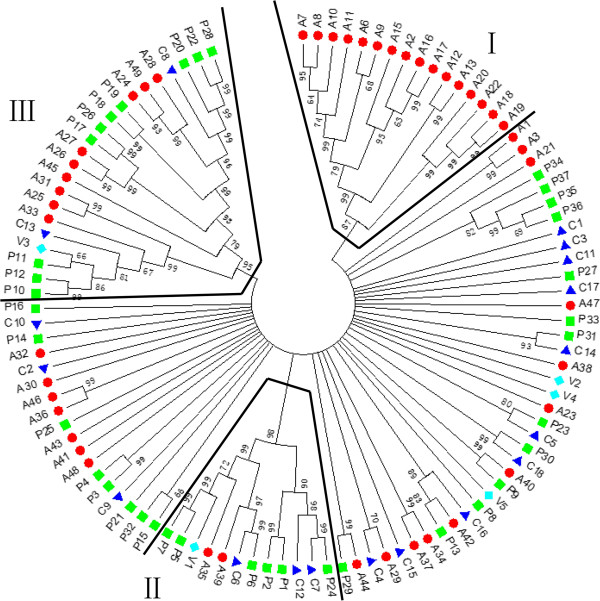
Phylogenetic tree of extensins constructed by the ME method with MEGA for the four modern rosids.
Next, we compared the phylogenetic trees described above with those generated using more advanced ML and BI methods. The ML tree constructed using the GTR+G model is shown in Additional file 5. The BI tree constructed using the GTR-CAT+G4 model is shown in Additional file 6. The topology of the ML tree was roughly consistent with that of the BI tree for branches with greater than 80% bootstrap support. When the ML and BI trees were compared with the ME and NJ trees at ≥60% bootstrapping support, the topology of the formers pair of trees differed significantly from that of the latter pair. Clades II and III, which were distinct in the ME and NJ trees, were collapsed in the ML and BI trees. However, when we separately combined the results for branches with more than 80% bootstrap support in the ME and NJ trees and did the same in the ML and BI trees, the combined results of the former were almost completely consistent with the combined results of the latter. The only exception was the emergence of A1 and A3, which was only supported with a bootstrap value ≥80% in the ME and NJ trees.
We further evaluated the phylogenetic relationship of paralogous and orthologous extensins in the four rosids (Table 1). We found that any of the paralogous and orthologous pairs were located in the same clade on the ME and NJ trees, suggesting the ME and NJ trees were ideal to infer the phylogenetic relationship of the paralogous and orthologous extensins with bootstrap support ≥60%.
Expansion manners of extensingenes
From the PGDD [32], we first retrieved the SCBs associated with the expansion of extensins within the genome of each species (Additional file 7). The number of extensins that arose from S/WGD varied among the four rosids. We detected 10, 21, two and two extensin genes associated with S/WGDs in Arabidopsis, Populus, Carica, and Vitis, respectively. Because the duplicated genes located on a SCB pair have duplicated simultaneously, the median Ks value of duplicated genes in SCBs can be used to infer the associated WGDs [5]. In Populus, the overall median Ks value of the duplicated genes related to the γ triplication event was 1.54, and that associated with the P-WGD was 0.27, as reported by Tang et al. [36]. We detected 36 SCB pairs associated with the expansion of extensin genes within the genome of Populus (Additional file 7). The median Ks of duplicated genes in different SCBs showed two distinct ranges (Figure 2a): 0.2437–0.3345, and 1.2633–1.7896. According to the calculations of Tang et al. [36], we proposed that SCBs with median Ks values in the range of 0.2437–0.3345 were associated with the P-WGD, and those with median Ks values in the range of 1.2633–1.7896 were likely related to the most ancient γ-triplication event (Figure 2a). For example, SCBs containing P5 and P7 likely arose from the P-WGD, as did the SCBs containing P6 and PN1. However, the median Ks value of any SCB combination (P5-PN1, P5-P6, P7-PN1, P6-P7) across the above two SCB pairs was in the range of 1.2633–1.7896. This finding suggested that the archetypal chromosomal blocks of these two SCB pairs (P5-P7, P6-PN1) were duplicates resulting from the γ-triplication event. The counterparts of extensins on many SCBs were found to have subfunctionalized into non-extensins (indicated by an “N” between the first letter and the number in the code name).
Figure 2.
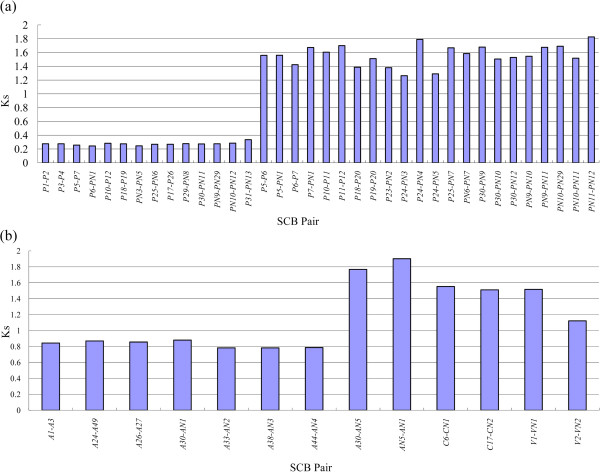
Median Ks values of SCB pairs associated with the expansion of extensins within the genomes of each species. a: Within the genome of Populus. b: Within the genomes of the other three species.
We further examined the SCBs associated with the expansion of extensins within the genomes of the other three species. In Arabidopsis, the median Ks values associated with β- and γ-WGDs were close to the saturation median Ks value of 2.00 [36]. Thus, the β- and γ-paleopolyploidies were indistinguishable based on the Ks value. The overall median Ks value associated with α-duplication was reported to be 0.86 [36]. In total, 10 extensin genes were associated with α-WGD in Arabidopsis, and the median Ks value of SCBs containing these genes were in the range of 0.783–0.881 (Figure 2b). Among these SCBs, the SCB pair A30-AN1 had a median Ks value of 0.881, suggesting this pair resulted from α-WGD. The SCB pair A30-AN5 had a median Ks value of 1.7678, suggesting that this pair was associated with β- or γ-paleopolyploidy. In both cases, the counterparts of extensins on the corresponding SCBs had subfunctionalized into non-extensins (AN1, AN5). In Carica and Vitis, the overall median Ks value of SCBs associated with γ triplication was 1.76 and 1.22, respectively [36]. In this study, we detected two SCBs containing extensin genes (C6-CN1, C17-CN2) in Carica; these pairs had median Ks values of 1.5526 and 1.5113, respectively (Figure 2b). This finding suggested that both duplicates resulted from γ-WGD. In Vitis, the SCB pair V1-VN1 had a median Ks value of 1.5164 (Figure 2b). The two chromosomal blocks of this SCB pair were located on Vitis chromosomes 1 and 14, and each had 37 duplicated genes in identical order. The other SCB pair, V2-VN2, had a median Ks value of 1.1221. The formation of the above two SCB pairs was associated with γ-WGD in Vitis.
In the above analyses, we analyzed the SCBs associated with the expansion of extensin genes within the genome of each species. Using the PGDD database [32], we also examined the SCBs associated with the expansion of extensins between species (Additional file 8). When a gene-collinearity analysis was conducted between species, we detected more extensins resulting from S/WGDs. For example, when the gene-collinearity analysis was conducted within a single species, we found that A33 and AN2 in Arabidopsis originated from α-WGD. When the gene-collinearity analysis was conducted between species, we found that the chromosomal blocks containing A33, AN2, A25, and A31 were all orthologous SCBs of the chromosomal block containing C13 in Carica. Therefore, these four genes originated from a common ancestral gene, and the paleopolyploidy associated with A25 and A31 should be more ancient than the α-WGD event. However, whether A25 and A31 resulted from β- or γ-WGD could not be determined because the median Ks values between β- and γ-SCBs in Arabidopsis were close to saturation. In Arabidopsis, extensins arising from α-WGD can be identified with certainty, but those resulting from β- and γ-WGD are undistinguishable. Similarly, the results of the gene-collinearity analysis between species revealed that A35 and A39 were associated with β- or γ-WGD in Arabidopsis.
Based on the gene-collinearity analysis within and between species, we established a panoramic picture of the differential retention and expansion of the ancestral extensins associated with paleopolyploidy in the four modern rosids (Figure 3). The retention and expansion of 24 ancestral extensins in these four modern rosids could be tracked unambiguously through gene collinearity analyses. The duplicates of these ancestral genes through S/WGDs were differentially retained in each species. We detected 19, 23, 6, and 4 extensin genes associated with paleopolyploidy in Arabidopsis, Populus, Carica, and Vitis, respectively (Figure 3). All of the other duplicates of these 24 ancestral genes that arose through S/WGDs were either subfunctionalized into non-extensins or completely lost (Figure 3). For instance, all genes shown in the first line of Figure 3 originated from the same ancestral gene (ancestral extensin-1). The genes that originated from this ancestral gene through ancient S/WGDs, including V1, C6, P5, P7, P6, A35 and A39, were remained as extensins; whereas the duplicates of this ancestral extensin, including VN1, CN1, PN1, AN21, AN22, and AN23 were subfunctionalized into non-extensins. Three duplicates of this ancestral extensin were lost from Arabidopsis, but the corresponding SCBs were retained (represented by an “L” at the corresponding position). Eight whole-SCBs have been completely lost from these four modern rosids (blank at the corresponding positions).
Figure 3.
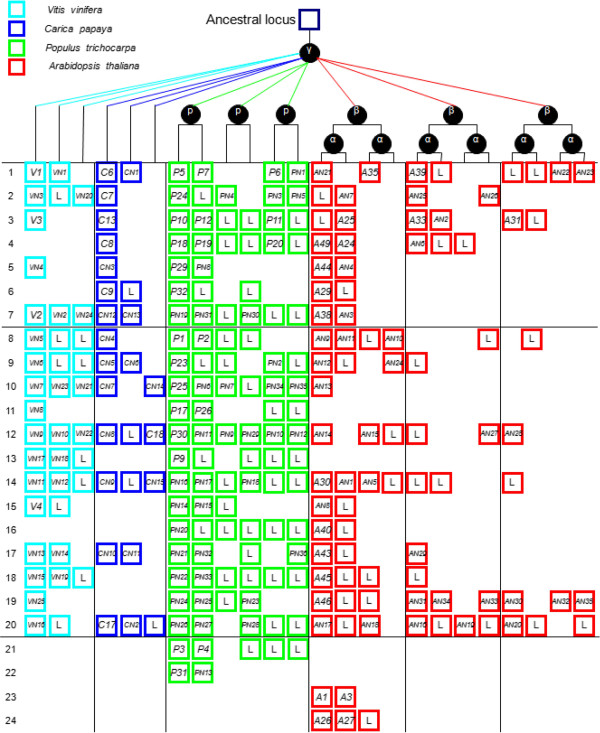
Panoramic picture to visualize the differential retention and expansion of the ancestral extensins associated with paleopolyploidy events that have occurred in four modern rosids. Notes: Square represents a SCB duplicated through paleopolyploidy events within and between species. Codes in the square correspond to associated extensin genes. Genes in the same line are thought to have originated from the same ancestral gene. Genes coded with a letter followed by a number (e.g., V1) represent genes retained as extensins; genes coded with an “N” between the letter and the number (e.g., CN1) represent those that have subfunctionalized into non-extensins; “L” represents duplicated extensin that has been lost, but the corresponding SCB has been retained; blank positions correspond to situations where the whole SCBs has been completely lost.
Besides extensins associated with paleopolyploidy, eight extensins in Arabidopsis were found to result from TDs, including A2, A17, A7, A8, A9, and A10 in the large paralogous group and another paralogous pair, A18-A19 (Table 1). In Populus, four extensins in the paralogous group (P34, P35, P36, and P37; Table 1) were generated through TDs; whereas in Carica and Vitis, no extensin genes had expanded through TD. Among the extensins expanded through TDs, none originated from the 24 ancestral extensins shown in Figure 3.
Regardless of the type of gene duplication event, all paralogous genes were likely duplicated from the same ancestral gene [37]. In Arabidopsis, we detected eight extensins (A6, A11, A12, A13, A15, A16, A20, and A28) that had expanded through duplication manners other than S/WGD or TD. In Populus, two paralogous extensins, P22 and P28, were identified to have duplicated via other manners. In Carica and Vitis, no paralogous extensins expanded via duplication manners other than S/WGD or TD. Among the extensins that expanded through other duplication manners, A28 (paralogs of A24 and A49, Table 1), P22 and P28 (paralog of P20, Table 1) originated from the same ancestral gene, one of the 24 ancestral extensins mentioned above (line 4, Figure 3).
The ortholog analysis revealed that C18 and P30 were an orthologous pair. A detailed examination revealed that C18 is located on a small contig in the C. papaya genome assembly [27]; this region contains only two genes and did not qualify for the gene collinearity analysis. C18 and P30 originated from the same ancestral gene (line 12, Figure 3).
Besides the extensins that expanded via traceable events, there were 11 other extensins unique to Arabidopsis (A21, A22, A23, A32, A34, A36, A37, A41, A42, A47, A48), eight unique to Populus (P8, P13, P14, P15, P16, P21, P27, P33), 11 unique to Carica (C1, C2, C3, C4, C5, C10, C11, C12, C14, C15, C16) and one unique to Vitis (V5). We propose that these extensins might represent the oldest relics of ancient extensins differentially retained in each species. Alternatively, some might be ancient intra-specific duplicates, but their paralog-ship was no longer traceable because of severe gene divergence or erosion of duplication signatures.
Large paralogous extensingroup in Arabidopsis
We detected a large paralogous group containing 12 extensins genes (A2, A6, A7, A8, A9, A10, A11, A12, A13, A15, A16, and A17) in Arabidopsis (Table 1). A paralog analysis suggested that all of these genes originated from a common ancestral gene. The ancestral extensin was retained and specifically expanded in the genome of Arabidopsis, but was lost from the genomes of the other three rosids. The remarkable and specific proliferation of this ancestral extensin in Arabidopsis suggests that this group of extensin genes plays an unusually important role in the biology of Arabidopsis. Using the method of Blanc and Wolfe [3], we reconstructed a tentative phylogeny of this large paralogous group (Figure 4). In Arabidopsis, the Ks values of paralogous pairs associated with the most recent α-WGD range of 0.72–0.99 [5].
Figure 4.
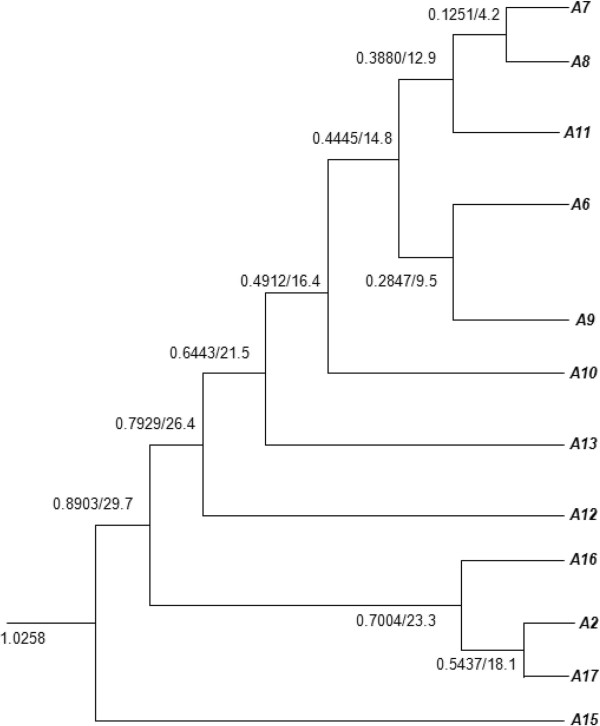
Tentative phylogeny constructed for a large paralogous group of extensins in Arabidopsis .
According to the Ks values shown in Figure 4, A15 is the most ancient extensin in this paralogous group, diverging from the other 11 extensins before the α-WGD. Around 29.7 MYs ago, A16, A2 and A17 further diverged from the remaining eight extensins. As shown in Figure 3, none of these extensins expanded through S/WGDs. Therefore, apart from A15, all of the other extensins in this paralogous group duplicated after the α-WGD. Among them, A2, A7, A8, A9, A10, and A17 expanded through TDs, while the others proliferated through other duplication manners. The genes in this paralogous group duplicated very actively. On average, duplication occurred around every 3 MYs, and the most recent duplication event occurred approximately 4 MYs ago between A7 and A8 through TD.
Discussion
A. thaliana, P. trichocarpa, C. papaya, and V. vinifera originate from a common paleohexaploid ancestor, and more recent paleopolyploidy events have recurred in the genomes of Populus and Arabidopsis. Paleopolyploidy events lead to gene duplication, which is believed to play a major role in evolutionary innovation [38]. Ancient genome duplications offer opportunities for the evolution of new genes [39] or genes with modified functions [40], changes in gene dosages, and the formation of new gene arrangements [36].
Relationships among genes in modern plants can be inferred based on sequence similarity, phylogenetic distance, and syntenic SCBs. However, each method has its own constraints. For instance, sequence similarity may have severely eroded during the evolutionary process for many of the ancient duplicates. Consequently, the paralog-ship for such duplicates would no longer be traceable based on sequence similarity. For example, our analyses showed that A1 and A3 are duplicates resulting from α-WGD in Arabidopsis, and P3 and P4 are duplicates that arose from P-WGD in Populus (Figure 3), but these paralogous pairs were not detected in the paralog analysis based on gene similarity (Table 1). More ancient duplicates could be tracked based on gene collinearity on SCBs within and between species. Yet, based on the gene collinearity analysis alone, we could not detect the paralogous pairs that arose via duplication manners other than S/WGDs (e.g., the large paralogous group unique to Arabidopsis; Table 1).
It is problematic to infer evolutionary relationships for homologous genes among different lineages based on phylogenetic distance alone. Gene variation rates vary among lineages. Therefore, inferring evolutionary relationships based on phylogenetic trees can produce incongruous results, because the drastic differences in rates may lead to incorrect trees that are artifacts because of long-branch attractions [41]. Therefore, to reconstruct the evolutionary relationships for members of a gene family across plant species, it is essential to combine all of the above analytical methods.
Based on gene collinearity on SCBs within and between species, we built a panoramic picture to display the differential retention and expansion of 24 ancestral extensins among the four modern rosids (Figure 3). Our analyses showed that seven (line 1–7) of these ancestral extensins were retained in more than two species, but only two (line 1 and line 3) were retained in all four modern rosids. By contrast, 17 of the ancestral extensins were retained in only one of the four rosids. Thus, most of the extensins in each modern rosid are descendants of different ancestral genes. This finding suggests that, using Arabidopsis as the model plant, we can only learn a limited amount about the functions of a gene family. If Arabidopsis is the only study material, we might not resolve the function of genes uniquely retained in the other rosids (e.g., V4 in Vitis, C17 in Carica, and P31 in Populus). Similarly, genes uniquely retained in Arabidopsis may have a specific and indispensable function in the species. For example, studies on A1 (Extensin-1) and A3 (Extensin-3) have demonstrated their importance in maintaining normal cell wall function in Arabidopsis [12, 14, 20, 42]. In particular, the RSH extensin (Extensin-3) was shown to play an essential role in the initiation of new cell growth [20]. Because A1 and A3 were uniquely retained in Arabidopsis, it remains unknown whether their function was compensated for by other extensins in the other three rosids, or whether such function was totally lost.
Considering the paleopolyploidy events that occurred in each species, there should be three ancestral loci in Carica and Vitis, because they only underwent the γ-triplication event. There should be six ancestral loci in Populus, because it underwent both γ-triplication and P-duplication, and 12 in Arabidopsis because it underwent γ-triplication, and then β- and α-duplications. However, such extreme values were not observed for any of the ancestral extensins. After the paleopolyploidy events, the exponential growth in gene numbers is often tempered by massive and progressive gene death in the subsequent diploidization process [4]. In this study, about 91.3% extensins associated with paleopolyploidies were found to have subfunctionalized into non-extensins or to have been completely lost from these four modern rosids (Figure 3). The convergent restoration of some genes to singleton status after multiple rounds of duplication in independent lineages suggests that there may be selective advantages for the organism to have only a single copy of these genes [43]. In most cases, we found that only one copy of the duplicated extensins resulting from ancient WGDs had been retained (Figure 3). In another study, such extensins were identified as “duplication resistant”; that is, only one copy per nucleus was adaptive [36].
As well as the γ-triplication event, Arabidopsis was affected by two more recent paleopolyploidy events (β- and α-duplication). In Populus, there was only one additional duplication (P-duplication), and the P-duplication in Populus was more ancient than the most recent α-duplication in Arabidopsis [5, 6]. However, Arabidopsis retained fewer extensins that proliferated through S/WGDs than did Populus. This may result from rapid substitutions in Arabidopsis. The γ-triplication event apparently occurred in the common ancestor of the four modern rosids. However, the median Ks between γ-paleologs in Arabidopsis (close to the saturation value of 2.00) was higher than that in Populus (1.54), Carica (1.76), and Vitis (1.22) [36]. This result suggests that more rapid substitutions occurred at synonymous sites in Arabidopsis than in the other three rosids. The high median Ks between γ- paleologs in Arabidopsis may be related to the more extensive chromosome rearrangements that occurred in Arabidopsis. There were three fusions, two translocations, and one inversion event during the 5 MYs after Arabidopsis lyrata diverged from A. thaliana [34]. This would give a rate estimate of 0.6 rearrangement/MY in the genome of these two species. The more extensive rearrangements of the chromosomal segments that occurred in Arabidopsis would destroy collinearity [44]. In species that have undergone several ancient WGDs, the more recent WGDs tend to obscure the collinearity from the more ancient ones [36]. Additionally, more SCBs were completely lost (blank sites in Figure 3) from Arabidopsis than from Populus. This result indicates that heavier contraction of chromosome blocks in Arabidopsis than in Populus accompanied the WGDs in the past. This idea is consistent with the fact that these two lineages originated from a common ancestor, and that Arabidopsis contains more paleopolyploidies, yet has a much smaller genome than that of Populus.
Besides the extensins expanded through S/WGDs, some extensins proliferated more actively through duplication manners other than S/WGD. Extensins in the latter category were more abundant in Arabidopsis and Populus than in Carica and Vitis. Among the genomes of the four rosids, that of Vitis shows the closest karyotype to that of their common ancestor [7]. Our analyses showed that almost all Vitis extensins are old relics of ancestral extensins from the common paleohexaploid ancestor (Figure 3). Both Carica and Vitis were only affected by the γ-WGD event, but Carica contains many more extensins than does Vitis. Among the Carica extensins, only seven show clear evidence of being relics of the common ancestor (Figure 3). Although we cannot exclude the possibility that some of the remaining extensins might represent ancient intra-specific duplicates that are no longer traceable, it is certain that there are no recently duplicated extensins in Carica. The fact that there are more extensins in Carica than in Vitis may imply that it uses more of these proteins in the development process and for adaptation to the environment.
A striking finding of this study is that extensins with the IPR006706 motif specifically expanded in A. thaliana. Such extensins were rare or completely absent from the other three rosids (Additional file 1). Both C. papaya and A. thaliana are in the Brassicaceae, yet C. papaya has only one extensin with the IPR006706 motif (C11). Selection after a duplication event contributes substantially to gene novelty, and hence, to functional divergence of genes in plants [45]. The ω values for paralogous extensins encoding proteins with the IPR006706 domain indicate that this group has been subjected to strong purifying selection. This finding highlights the importance of the function of this highly conserved gene group in Arabidopsis.
Referring to the phylogenetic tree at the phytozome website (http://www.phytozome.net/alyrata.php), we further analyzed the expansion of such extensins in close relatives of A. thaliana, including A. lyrata, Capsella rubella, and Brassica rapa. All of these species are members of the Brassicaceae. Similar to the case in A. thaliana, such extensins had also significantly expanded in these lineages. We detected 10, 7, and 15 extensins with an IPR006706 motif in A. lyrata, C. rubella, and B. rapa, respectively. Like in Arabidopsis, more than 50% of these extensins had expanded through TDs in each species. Additionally, nine, six, three, and six of these extensins were shared orthologs between species in A. thaliana, A. lyrata, C. rubella and B. rapa, respectively. Thus, the expansion of this extensins group occurred after the progenitor of the above four lineages diverged from Carica.
Conclusions
Based on sequence similarity, phylogenetic distance and gene collinearity on the SCBs, we tracked the differential retention and expansion of ancestral extensin genes among four modern rosids. The results revealed that most of the extensins in each species are descendants of different ancestral genes. We also detected a group of extensins that specifically expanded in the Arabidopsis genome. An important group of extensins has been retained only in Arabidopsis. Whether their function was compensated for by other extensins or such function was totally lost in the other rosids remains unknown. These findings highlight that we can only learn a limited amount about the functions of a particular gene family using Arabidopsis as the model plant. This study also highlights the importance of learning the origin of a gene when analyzing its function across different plant species.
Availability of supporting data
The data set(s) supporting the results of this article hyperlink to dataset(s) in http://www.arabidopsis.org (TAIR10 release of November 2010), ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Ptrichocarpa/ (Populus trichocarpa), ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Vvinifera (Vitis vinifera), and ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Cpapaya/ (Carica papaya).
The relevant primary data have been deposited in TreeBase, with Study Accession URL: http://purl.org/phylo/treebase/phylows/study/TB2:S16026.
Authors’ information
Lianhua Guo: Ph.D Nanjing Forestry University. He is now a faculty member at Zhejiang Agriculture & Forestry University.
Yingnan Chen: Ph.D, faculty member of Nanjing Forestry University.
Ning Ye: Ph. D, faculty member of Nanjing Forestry University.
Xiaogang Dai: Ph.D, Nanjing Forestry University.
Wanxu Yang: Graduate student at Nanjing Forestry University.
Tongming Yin: Ph.D, laboratory director and full time professor at Nanjing Forestry University.
Electronic supplementary material
Additional file 1: Extensins identified in four modern rosids. (XLS 38 KB)
Additional file 2: Number of extensins with different signature protein domains in four modern rosids. (XLS 30 KB)
Additional file 3: ω values for the paralogous and orthologous pairs of extensins. (XLS 34 KB)
Additional file 4: Phylogenetic tree of extensins constructed by NJ method with MEGA for four modern rosids. (JPEG 719 KB)
Additional file 5: Phylogenetic tree of extensins constructed by ML method with GTR+G model. (JPEG 312 KB)
Additional file 6: Phylogenetic tree of extensins constructed by BI method with GTR-CAT+G4 model. (JPEG 278 KB)
Additional file 7: Extensins in syntenic chromosomal blocks identified within each species. (XLSX 39 KB)
Additional file 8: Extensins in syntenic chromosomal blocks identified between species. (XLS 49 KB)
Acknowledgments
This work was funded by the Key Forestry Public Welfare Project (201304102), the National Basic Research project (2012CB114505), the Natural Science Foundation (31125008), the Program for Innovative Research Team in Universities of Jiangsu Province and the Educational Department of China. This research was also supported by the Doctorate Fellowship Foundation and the Priority Academic Program Development (PAPD) Program at Nanjing Forestry University.
Abbreviations
- PGDD
The plant genome duplication database
- S/WGD
Segmental or whole-genome duplications
- SCBs
Syntenic chromosomal blocks
- TD
Tandem duplication.
Footnotes
Competing interests
The authors’ declare that they have no competing interests.
Authors’ contributions
LG and YC conducted the data collection, and participated in data analysis and drafting the manuscript. NY participated in the gene duplication analysis. XD participated in the phylogenetic study in the statistical analysis. WY participated in the extensins identification and sequence alignment. TY conceived of the study and participated in the design and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Lianhua Guo, Email: glh@zafu.edu.cn.
Yingnan Chen, Email: chenyingnan@njfu.edu.cn.
Ning Ye, Email: yening2004@gmail.com.
Xiaogang Dai, Email: xgdai@njfu.edu.cn.
Wanxu Yang, Email: yangwx394@126.com.
Tongming Yin, Email: tmyin@njfu.com.cn.
References
- 1.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 2.Adams KL, Percifield R, Wendel JF. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics. 2004;168:2217–2226. doi: 10.1534/genetics.104.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 5.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, et al. The genome of black cottonwood, Populus trichocarpa. Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 7.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 8.Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010;153:485–513. doi: 10.1104/pp.110.156554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tierney ML, Varner JE. Review the extensins. Plant Physiol. 1987;84:1–2. doi: 10.1104/pp.84.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieliszewski M, Lamport DT. Extensin: repetitive motifs, functions sites, post-translational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313X.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- 11.Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Fachin LP, Egelund J, Gille S, Harholt J, Ciancia M, Verli H, Pauly M, Bacic A, Olsen CE, Ulvskov P, Petersen BL, Somerville C, Iusem ND, Estevez JM. O-glycosylated cell wall proteins are essential in root hair growth. Science. 2011;332:1401–1403. doi: 10.1126/science.1206657. [DOI] [PubMed] [Google Scholar]
- 12.Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DT, Chen Y, Kieliszewski MJ. Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci U S A. 2008;105:2226–2231. doi: 10.1073/pnas.0711980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamport DTA, Kieliszewski MJ, Che Y, Cannon MC. Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 2011;156:1–25. doi: 10.1104/pp.111.900410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts K, Shirsat AH. Increased extensin levels in Arabidopsis affect inflorescence stem thickening and height. J Exp Bot. 2006;57:537–545. doi: 10.1093/jxb/erj036. [DOI] [PubMed] [Google Scholar]
- 15.Goldman MH, Pezzotti M, Seurinck J, Mariani C. Developmental expression of tobacco pistil-specific genes encoding novel extensin-like proteins. Plant Cell. 1992;4:1041–1051. doi: 10.1105/tpc.4.9.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinstein AL, Marquez J, Suarez-Cervera M, Bedinger PA. Extensin-like glycoproteins in the maize pollen tube wall. Plant Cell. 1995;7:2211–2225. doi: 10.1105/tpc.7.12.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Ren Y, Zhao J. Roles of extensins in cotyledon primordium formation and shoot apical meristem activity in Nicotiana tabacum. J Exp Bot. 2008;59:4045–4058. doi: 10.1093/jxb/ern245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkouropoulos G, Barnett DC, Shirsat AH. The Arabidopsis extensin gene is developmentally regulated, is induced by wounding, methyl jasmonate, abscisic and salicylic acid, and codes for a protein with unusual motifs. Planta. 1999;208:212–219. doi: 10.1007/s004250050552. [DOI] [PubMed] [Google Scholar]
- 20.Hall Q, Cannon MC. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell. 2002;14:1161–1172. doi: 10.1105/tpc.010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, Courcelle E, Das U, Daugherty L, Dibley M, Finn R, Fleischmann W, Gough J, Haft D, Hulo N, Hunter S, Kahn D, Kanapin A, Kejariwal A, Labarga A, Langendijk-Genevaux PS, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Kalluri UC, Jawdy S, Gunter LE, Yin T, Tschaplinski TJ, Weston DJ, Ranjan P, Tuskan GA. The F-Box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 2008;148:1189–1200. doi: 10.1104/pp.108.121921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden LT, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Reserch. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Kuma K, Miyata T, Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform Series. 2005;16:22–33. [PubMed] [Google Scholar]
- 25.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia X, Xie Z. DAMBE: data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 32.Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41:1152–1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang SG, Kim DS, Jang CS. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica. 2011;139:1023–1032. doi: 10.1007/s10709-011-9604-y. [DOI] [PubMed] [Google Scholar]
- 34.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galbo T, Olsen GS, Quistorff B, Nishimura E. Free fatty acid-induced PP2A hyperactivity selectively impairs hepatic insulin action on glucose metabolism. PLoS ONE. 2011;6:e27424. doi: 10.1371/journal.pone.0027424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohno S. Evolution by Gene Duplication. New York, American: Springer-Verlag Press; 1970. [Google Scholar]
- 39.Spillane C, Schmid KJ, Laoueille-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U. Positive Darwinian selection at the imprinted MEDEA locus in plants. Nature. 2007;448:349–352. doi: 10.1038/nature05984. [DOI] [PubMed] [Google Scholar]
- 40.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 41.Felsenstein J. Inferring Phylogenies. Sunderland, USA: Sinauer Associates press; 2004. [Google Scholar]
- 42.Wei G, Shirsat AH. Extensin over-expression in Arabidopsis limits pathogen invasiveness. Mol Plant Pathol. 2006;7:579–592. doi: 10.1111/j.1364-3703.2006.00363.x. [DOI] [PubMed] [Google Scholar]
- 43.Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006;22:597–602. doi: 10.1016/j.tig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Kuittinen H, de Haan AA, Vogl C, Oikarinen S, Leppala J, Koch M, Mitchell-Olds T, Langley CH, Savolainen O. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics. 2004;168:1575–1584. doi: 10.1534/genetics.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gossmann TI, Schmid KJ. Selection-driven divergence after gene duplication in Arabidopsis thaliana. J Mol Evol. 2011;73:153–165. doi: 10.1007/s00239-011-9463-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Extensins identified in four modern rosids. (XLS 38 KB)
Additional file 2: Number of extensins with different signature protein domains in four modern rosids. (XLS 30 KB)
Additional file 3: ω values for the paralogous and orthologous pairs of extensins. (XLS 34 KB)
Additional file 4: Phylogenetic tree of extensins constructed by NJ method with MEGA for four modern rosids. (JPEG 719 KB)
Additional file 5: Phylogenetic tree of extensins constructed by ML method with GTR+G model. (JPEG 312 KB)
Additional file 6: Phylogenetic tree of extensins constructed by BI method with GTR-CAT+G4 model. (JPEG 278 KB)
Additional file 7: Extensins in syntenic chromosomal blocks identified within each species. (XLSX 39 KB)
Additional file 8: Extensins in syntenic chromosomal blocks identified between species. (XLS 49 KB)


