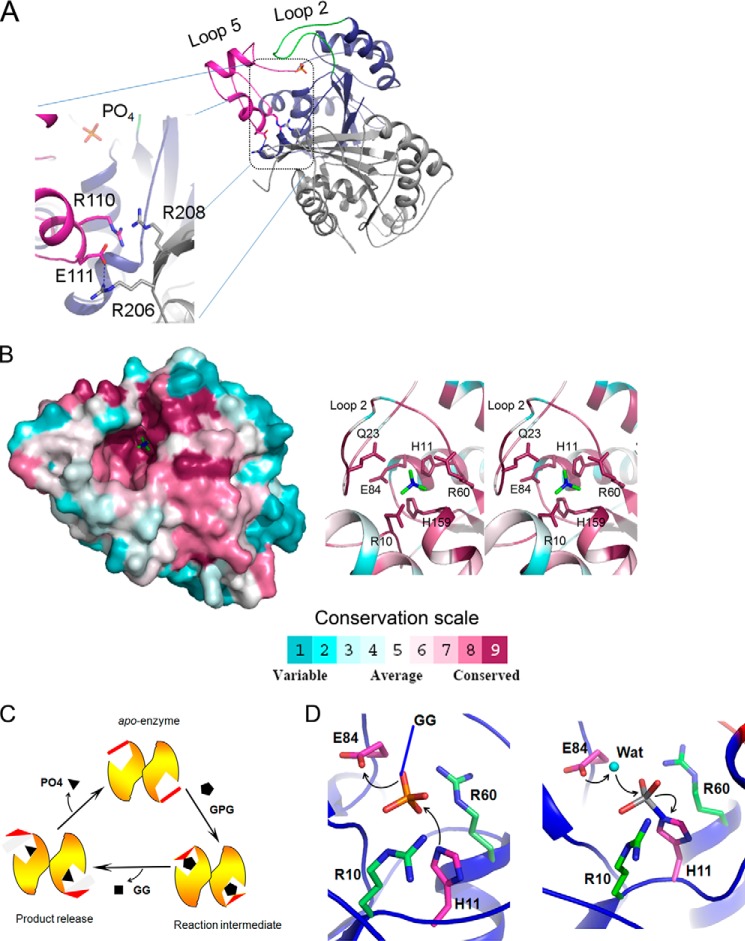
FIGURE 5.
Mechanism of catalysis. A, intermolecular ionic interactions help stabilize conformation of loop 5 during catalysis. Within a dimer of MtbGpgP, one monomer is colored blue, whereas the other is colored gray. Loop 2 and loop 5 of monomer 1 are colored in green and magenta, respectively. Residues involved in interactions and orthophosphate are shown as sticks. The hydrogen bond is shown as a dashed line. B, location of conserved residues (left panel) based on ConSurf analysis of primary sequence of 151 homologues of MtbGpgP (38). The surface of MtbGpgP is colored according to conservation depicted in scale. Residues around phosphate (green sticks) are highly conserved. The right panel shows a stereo view of close up of some of the conserved residues (sticks) around the phosphate (green sticks). C, diagrammatic representation of conformational changes of MtbGpgP during catalysis. Key conformational changes of loop 2 (red) during the conversion of GPG to GG have been captured in the crystal structures of MtbGpgP. D, catalysis proceeds in two steps. In step 1 (left panel), His11 (magenta sticks) mounts a nucleophilic attack on P center of phosphate (sticks). Glu84 (magenta sticks) donates a proton to the substrate. The substrate (GPG) is depicted as phosphate (sticks) linked to GG by a covalent bond shown in blue. In step 2 (right panel), a water molecule (cyan sphere) activated by Glu84 mounts an attack on the P center of the reaction intermediate. Amino acids stabilizing reaction intermediates are shown as green sticks. For clarity, Asn17, Gln23, and His159 participating in the stabilization are not shown.