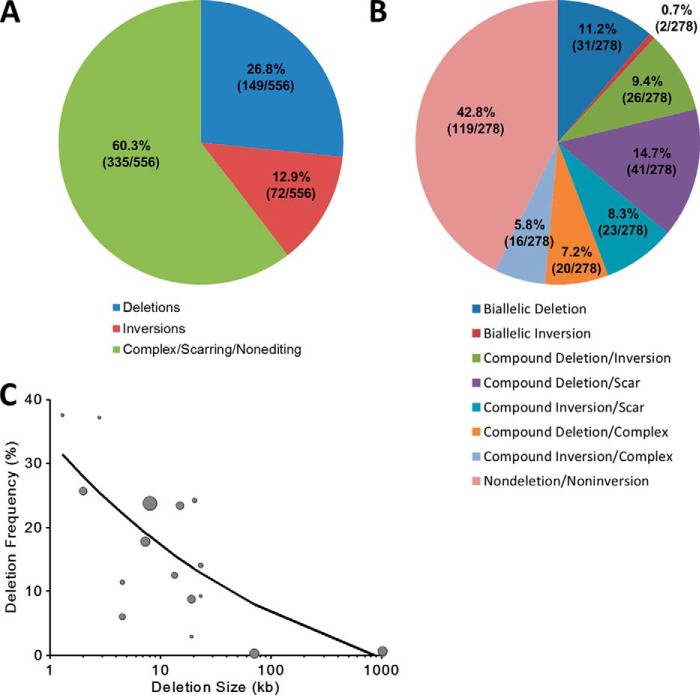
FIGURE 7.
Deletion/inversion frequency and characterization of outcomes when using two sgRNAs. A, deletion and inversion frequency were calculated on a per allele basis for all 278 clones (556 alleles) across the four loci examined in detail. B, 278 clones from across the four loci examined in detail were classified into eight categories based on the presence of deletion, inversion, scar, and complex alleles. C, deletion frequency inversely related to deletion size. Genomic deletion sizes ranged from 1.3 to 1,026 kb, which demonstrate a decrease in the frequency of deletion events as deletion size increases. The size of the circles corresponds to the number of clones screened for the corresponding sgRNA pair. The best fit relationship was determined by a weighted (by number of clones screened) nonlinear regression of the form function(deletion size) = k1 + k2(deletion size in kb)−k3, where k1, k2, and k3 represent constants: function(deletion size in kb) = −2.84 + 41.41(deletion size in kb)−0.36; R2 = 0.62. The weighted nonlinear regression was computed using the fitnlm function available in MATLAB R2013b software (MathWorks, Natick, MA).