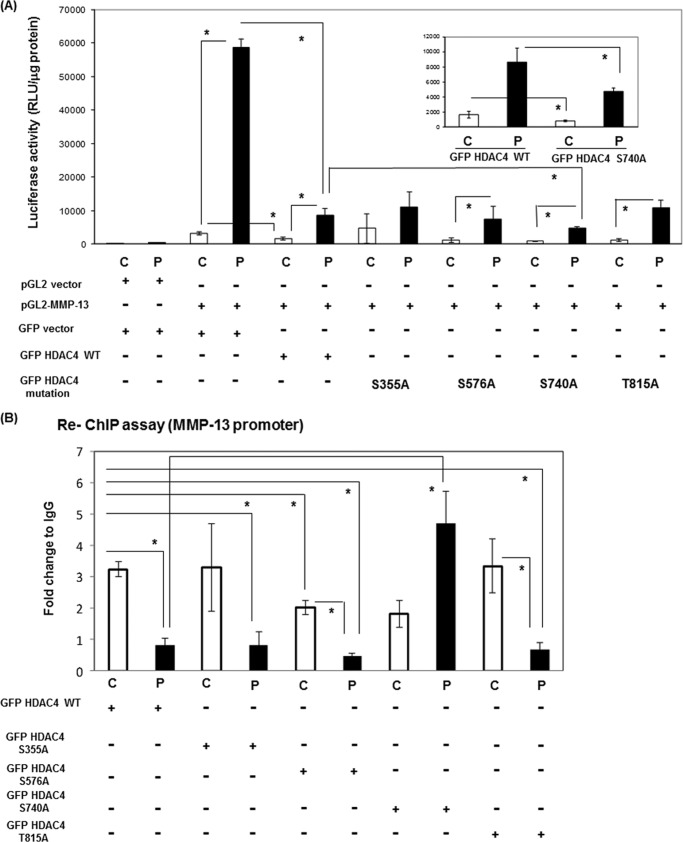
FIGURE 4.
GFP-rHDAC4-S740A mutation abolished dissociation from Runx2 on the MMP-13 promoter. A, UMR 106–01 cells were transiently transfected with various vectors (100 ng of pGL2 vector, 100 ng of -148 rat MMP-13 promoter-Luc, 100 ng of GFP vector, 100 ng of GFP rHDAC4, and mutant GFP rHDAC4 constructs), and the luciferase activities were measured with (P) or without (C) 6 h of 10−8 m PTH (1–34) treatment. The luciferase activities were normalized to total protein. Error bars represent ± S.E. of three independent experiments. *, p < 0.05 versus respective control. B, UMR 106-01 cells were transfected with GFP rHDAC4 or mutant GFP rHDAC4 constructs (S355A, S576A, S740A, and T815A). The cells were treated with control or PTH (10−8 m) for 30 min, then prepared for ChIP assays. After immunoprecipitation of the cross-linked lysates with anti-Runx2 or with rabbit IgG as a negative control, the immune complexes were collected then diluted with ChIP dilution buffer and re-subjected to ChIP assays with anti-GFP or rabbit IgG. The DNA was subjected to PCR with primers that amplify the distal RD and proximal AP-1 sites of the endogenous rat MMP-13 promoter. Input DNA (1:100) is a positive control for the assay. Data are shown as -fold change to control after normalizing to input and IgG. Error bars represent ± S.E. of three independent experiments. *, p < 0.05 versus respective control.