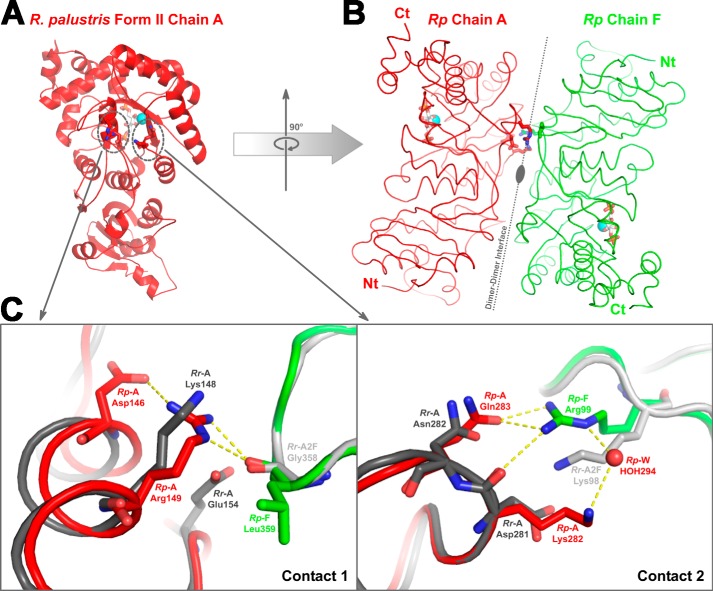
FIGURE 10.
Dimer-dimer interaction regions in R. palustris form II Rubisco (red and green) and the analogous region from R. rubrum form II (Protein Data Bank code 9RUB; dark and light gray) Rubisco. A, location of two groups of contact residues (dotted circles), which facilitate the association of dimer units via hydrogen bonds, as shown in one of the subunits (Rp Chain A; red ribbon) from the R. palustris enzyme. B, the association with neighboring subunit (Rp Chain F; green ribbon) is shown with an imaginary axis passing through the interface. Identical hydrogen bond contacts are present on the lower side of the dimer-dimer interface as well but are not shown in the figure. C, a close-up view of the two regions of contact aligned with the analogous region from R. rubrum Rubisco. Chain A from R. rubrum (Rr-A; dark gray) and R. palustris (Rp-A; red) enzymes were aligned, and chain A from another molecule of R. rubrum enzyme (Rr-A2F; light gray) was aligned to chain F of the R. palustris Rubisco (Rp-F; green). The hydrogen bond interactions present in the R. palustris structure are shown, and it is perceivable that similar interactions cannot be formed with analogous residues from the superimposed R. rubrum subunits. Ct, carboxyl terminus; Nt, amino terminus.