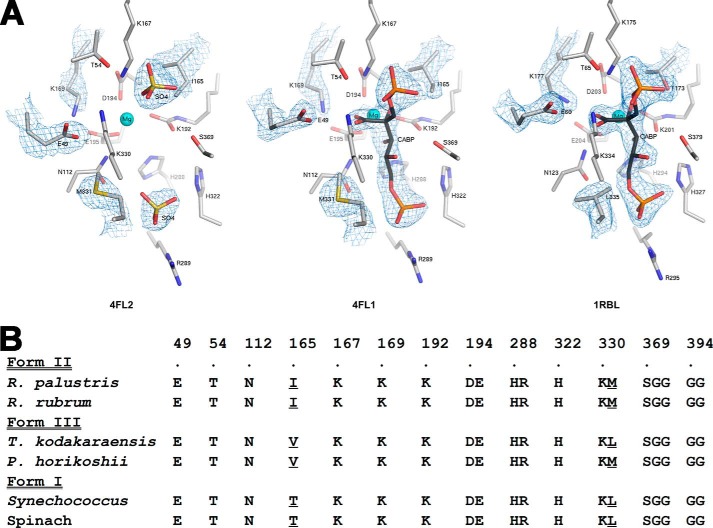
FIGURE 5.
Comparison of residues within 4 Å of CABP in several Rubisco structures. A, active site arrangements in the x-ray crystal structures of sulfate-bound (Protein Data Bank code 4LF2) and CABP-bound (Protein Data Bank code 4LF1) R. palustris form II Rubisco and CABP-bound Synechococcus form I Rubisco (Protein Data Bank code 1RBL). Electron density maps contoured between 1.0 and 2.0 σ for optimal visualization are displayed for the active site ligands and amino acid side chains with dissimilar conformations or identities. B, phylogenetic comparison of amino acid identities in this region. Sequences shown are representatives from the three forms of Rubisco. Residues appearing at different positions in the amino acid sequence are segregated into different columns, and the R. palustris form II Rubisco sequence numbers are indicated above the alignment along with a dot, which is aligned with the corresponding residue labels. Multiple residues within a column denote residues that are continuous in the amino acid sequence, and the sequence numbers correspond to those of the first residue within a column. Divergent residues chosen for analysis in this study and the analogous residues from other Rubisco sequences are underlined.