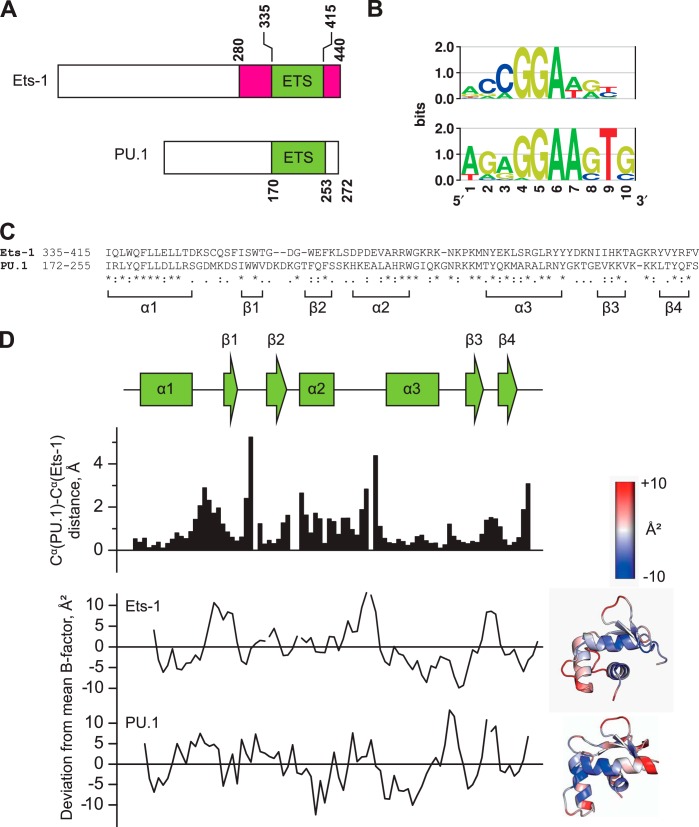
FIGURE 1.
The sequence-divergent ETS domains of PU.1 and Ets-1 adopt homologous backbone conformations. A, domain structures of murine Ets-1 and PU.1. The ETS domains of mouse and human counterparts are identical in sequence. The autoinhibitory flanking regions in Ets-1 are colored in magenta. B, sequence preferences of human Ets-1 and PU.1 as determined by ChIP sequencing and presented as relative entropy-weighted DNA logos (68). Positions at which only a single base is found contribute 2 bits of information content and positions at which all bases are equiprobable contribute 0 bits. C, sequence alignment of the ETS domains of Ets-1 and PU.1. Asterisks (*), colons (:), and periods (.) represent identity, conservative, and semiconservative differences, respectively. D, structural alignment of the high affinity protein·DNA complexes formed by the minimal ETS domains of Ets-1 (Protein Data Bank code 1K79) and PU.1 (Protein Data Bank code 1PUE). Scalar distances between aligned Cα and the deviation from the mean B-factor over the entire domain (37 and 16 Å2 for Ets-1 and PU.1, respective) are shown. The B-factor deviations are also spatially annotated in cartoon structures of the two ETS domains. Note that alignment is lowest at segments adjoining secondary structure elements, but these segments are not disordered as judged by their B-factors.