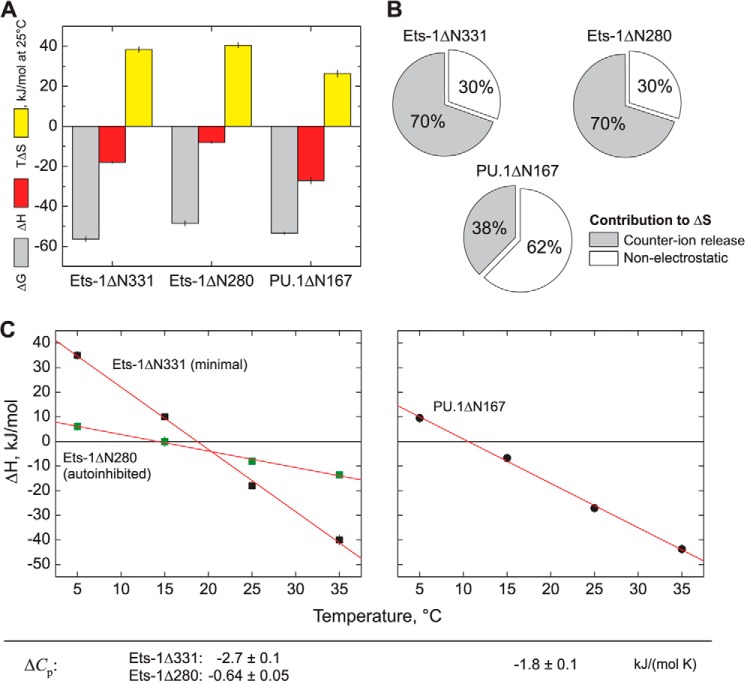
FIGURE 4.
Thermodynamic dissection of high affinity DNA site recognition by the ETS domains of Ets-1 and PU.1. A, comparative thermodynamics for minimal Ets-1ΔN331, autoinhibited Ets-1ΔN280, and PU.1ΔN167 at 25 °C, 150 mm Na+, and normal osmolality. Free energy values are derived from equilibrium affinity measurements: ΔG = −RT ln KB where R is the gas constant. Enthalpy changes for PU.1 are extracted from calorimetric measurements as previously described (29). Entropic contributions are computed as −TΔS = ΔG − ΔH. B, electrostatic contribution to the observed entropy changes. The entropy change caused by counter ion release is calculated from the salt dependence data (Fig. 3) as: ΔSCR = −ΨZR ln [M+] (26). C, temperature dependence of the enthalpy change of binding, yielding heat capacity changes (ΔCp) as the slope.