Extended Data Figure 5. Slx4/FancP depletion suppresses Tus/Ter-induced HR.
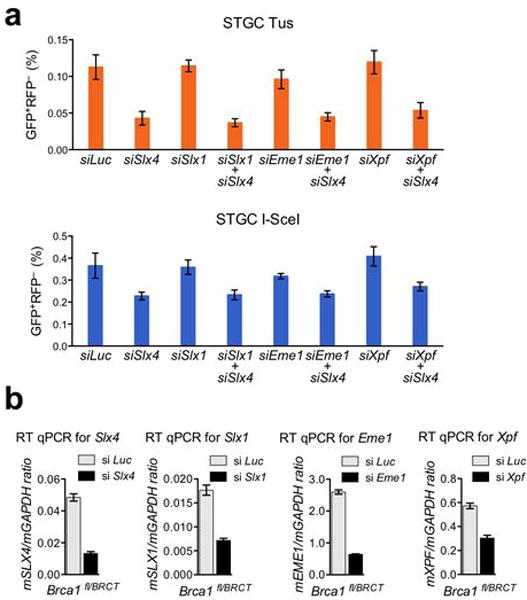
a, Frequencies of STGC in Brca1fl/BRCT 6xTer-HR cells co-transfected with Tus (orange) or I-SceI (blue) and with either control Luciferase siRNA (si Luc), Slx4 SMARTpool (si Slx4), Slx1 SMARTpool (si Slx1), Slx1 and Slx4 SMARTpools (si Slx1 si Slx4), Eme1 SMARTpool (si Eme1), Eme1 and Slx4 SMARTpools (si Eme1 si Slx4), Xpf SMARTpool (si Xpf), Xpf and Slx4 SMARTpools (si Xpf si Slx4). Each column represents the mean of triplicate samples from four independent experiments for each clone (i.e., n=4). Error bars: s.e.m. Tus-induced HR: t-test of si Slx4 vs. si Luc: P= 0.0219; si Slx4 vs. si Slx1: P= 0.0012; si Slx4 vs. si Slx4+1: P= 0.5983; si Slx4 vs. si Eme1: P= 0.0171; si Slx4 vs. si Slx4 +si Eme1: P= 0.8721; si Slx4 vs. si Xpf: P= 0.0098; si Slx4 vs. si Slx4+ si Xpf: P= 0.4711; si Slx1 vs. si Luc: P= 0.9332; si Eme1 vs. si Luc: P= 0.4631; si Xpf vs. si Luc: P= 0.7818; si Slx4+1 vs. si Luc: P= 0.0155; si Slx4+si Eme1 vs. si Luc: P= 0.0215; si Slx4 + si Xpf vs. si Luc: P= 0.0305. I-SceI-induced HR: t-test of si Slx4 vs. si Luc: P= 0.0907; si Slx4 vs. si Slx1: P= 0.0195; si Slx4 vs. si Slx4+1: P= 0.4897; si Slx4 vs. si Eme1: P= 0.0568; si Slx4 vs. si Slx4 +si Eme1: P= 0.3411; si Slx4 vs. si Xpf: P= 0.0745; si Slx4 vs. si Slx4+ si Xpf: P= 0.2726; si Slx1 vs. si Luc: P= 0.9198; si Eme1 vs. si Luc: P= 0.3349; si Xpf vs. si Luc: P= 0.9217; si Slx4+1 vs. si Luc: P= 0.1521; si Slx4+si Eme1 vs. si Luc: P= 0.2864; si Slx4 + si Xpf vs. si Luc: P= 0.2063. b, RT qPCR analysis of mRNA exon boundaries for Slx4, Slx1, Eme1, and Xpf mRNA in siRNA-SMARTpool-treated cells used in panel (a).
