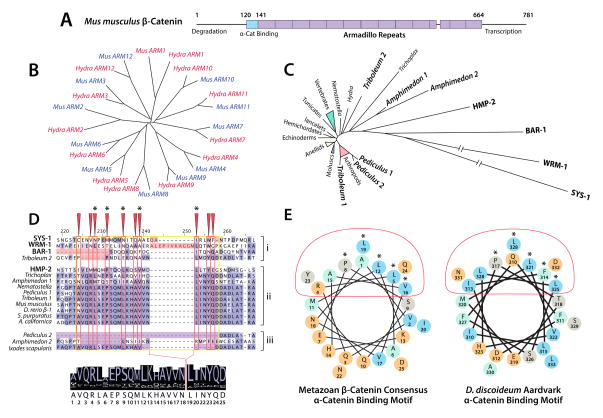
Figure 6. Evolutionary Constraint of β-Catenin by the α/β-Catenin Interaction.
(A) A schematic representation of Mm β-catenin, indicating the position of the 〈-catenin binding region and Armadillo repeats.
(B) A phylogeny of Hydra and Mus Armadillo repeats 1–12 (after (Schneider et al., 2003)) indicating homology of each repeat.
(C) A neighbor-joining phylogeny of metazoan β-catenins, indicating divergent paralogs in bold. Branch length correlates to number of amino acid substitutions between proteins.
(D) An alignment generated using MUSCLE (Edgar, 2004), visualized in JalView (Waterhouse et al., 2009), highlighting the α-catenin binding motif (yellow box), conserved binding surface (red boxes/arrows), and structurally important residues (asterisks). Paralogs that fail to bind α-catenin (i) and untested paralogs and orthologs with abberant binding motifs (iii) are segregated from α-catenin-binding paralogs and single orthologs (ii) for clarity. A consensus sequence generated from the proteins in group ii is displayed below.
(E) Helical wheel representations of the consensus α-catenin binding helix from D, and that of D. discoideum (after (Dickinson et al., 2011a)). Charged residues are colored orange, and hydrophobic residues are colored cyan. Red ovals and asterisks indicate the α-catenin binding surface and structurally important residues as in D.