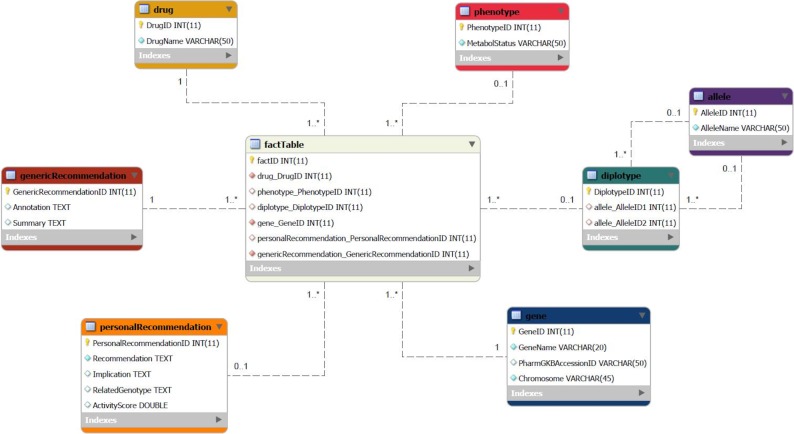
Figure 4.
The basic star-schema data model of the electronic PGx assistant: gene—any gene that is known to be associated with drug metabolism; defining attributes: GeneName, Chromosome and PharmGKB code (PharmGKBAccessionID); drug—any PGx drug; defining attributes: DrugName and PharmGKB code (PharmGKBAccessionID); factTable—the table that joins genes, drugs, diplotypes, phenotypes and recommendations; its primary (composite) key is a combination of the foreign keys of other tables; diplotype—the diplotype as defined by the two allele combination (i.e. *1/*2); defining attributes: the two allele IDs (Allele1ID and Allele2ID); allele—any star-allele haplotype (i.e. *1, *2, *1B, etc.); defining attribute: (AlleleName); genericRecommendation—a recommendation for a specific gene–drug combination; defining attributes: Summary and Annotation; personalRecommendation—specific recommendations when diplotype is known; defining attributes: Recommendation, Implication, RelatedGenotype (i.e. an individual carrying two loss-of-function alleles) and ActivityScore; phenotype—the different types of Metabolizer status (i.e. Intermediate Metabolizer, Extensive Metabolizer, etc.); defining attribute: MetabolStatus.